Conservation genetics in Chinese sheep: diversity of fourteen indigenous sheep (Ovis aries) using microsatellite markers
- PMID: 26865968
- PMCID: PMC4739567
- DOI: 10.1002/ece3.1891
Conservation genetics in Chinese sheep: diversity of fourteen indigenous sheep (Ovis aries) using microsatellite markers
Abstract
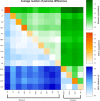
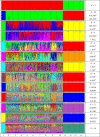
The domestic sheep (Ovis aries) has been an economically and culturally important farm animal species since its domestication around the world. A wide array of sheep breeds with abundant phenotypic diversity exists including domestication and selection as well as the indigenous breeds may harbor specific features as a result of adaptation to their environment. The objective of this study was to investigate the population structure of indigenous sheep in a large geographic location of the Chinese mainland. Six microsatellites were genotyped for 611 individuals from 14 populations. The mean number of alleles (±SD) ranged from 7.00 ± 3.69 in Gangba sheep to 10.50 ± 4.23 in Tibetan sheep. The observed heterozygote frequency (±SD) within a population ranged from 0.58 ± 0.03 in Gangba sheep to 0.71 ± 0.03 in Zazakh sheep and Minxian black fur sheep. In addition, there was a low pairwise difference among the Minxian black fur sheep, Mongolian sheep, Gansu alpine merino, and Lanzhou fat-tailed sheep. Bayesian analysis with the program STRUCTURE showed support for 3 clusters, revealing a vague genetic clustering pattern with geographic location. The results of the current study inferred high genetic diversity within these native sheep in the Chinese mainland.
Keywords: China; Diversity; indigenous sheep; microsatellite.
Figures
Similar articles
-
Genetic variation of 5 SNPs of MC1R gene in Chinese indigenous sheep breeds.Genetika. 2014 Oct;50(10):1188-99. doi: 10.7868/s0016675814100166. Genetika. 2014. PMID: 25720251
-
[Origin and genetic diversity of Mongolian and Chinese sheep using mitochondrial DNA D-loop sequences].Yi Chuan Xue Bao. 2005 Dec;32(12):1256-65. Yi Chuan Xue Bao. 2005. PMID: 16459654 Chinese.
-
Current genetic diversity in eight local Chinese sheep populations.Mol Biol Rep. 2019 Feb;46(1):1307-1311. doi: 10.1007/s11033-018-4445-8. Epub 2018 Dec 17. Mol Biol Rep. 2019. PMID: 30560407
-
Meta-Analysis of Mitochondrial DNA Control Region Diversity to Shed Light on Phylogenetic Relationship and Demographic History of African Sheep (Ovis aries) Breeds.Biology (Basel). 2021 Aug 10;10(8):762. doi: 10.3390/biology10080762. Biology (Basel). 2021. PMID: 34439994 Free PMC article. Review.
-
Trends towards revealing the genetic architecture of sheep tail patterning: Promising genes and investigatory pathways.Anim Genet. 2021 Dec;52(6):799-812. doi: 10.1111/age.13133. Epub 2021 Sep 1. Anim Genet. 2021. PMID: 34472112 Review.
Cited by
-
Genetic diversity of the Chinese goat in the littoral zone of the Yangtze River as assessed by microsatellite and mtDNA.Ecol Evol. 2018 Apr 24;8(10):5111-5123. doi: 10.1002/ece3.4100. eCollection 2018 May. Ecol Evol. 2018. PMID: 29876086 Free PMC article.
-
Genetic diversity of different breeds of Kazakh sheep using microsatellite analysis.Arch Anim Breed. 2019 Jun 5;62(1):305-312. doi: 10.5194/aab-62-305-2019. eCollection 2019. Arch Anim Breed. 2019. PMID: 31807641 Free PMC article.
-
Whole-genome resequencing to investigate the genetic diversity and mechanisms of plateau adaptation in Tibetan sheep.J Anim Sci Biotechnol. 2024 Dec 6;15(1):164. doi: 10.1186/s40104-024-01125-1. J Anim Sci Biotechnol. 2024. PMID: 39639384 Free PMC article.
-
Genetic diversity and phylogenetic relationship of nine sheep populations based on microsatellite markers.Arch Anim Breed. 2021 Jan 6;64(1):7-16. doi: 10.5194/aab-64-7-2021. eCollection 2021. Arch Anim Breed. 2021. PMID: 34084899 Free PMC article.
References
-
- Alvarez, I. , Gutiérrez J. P., Royo L. J., Fernández I., Gómez E., Arranz J. J., et al. 2005. Testing the usefulness of the molecular coancestry information to assess genetic relationships in livestock using a set of Spanish sheep breeds. J. Anim. Sci. 83:737–744. - PubMed
-
- Arora, R. , Bhatia S., Mishra B. P., and Joshi B. K.. 2011. Population structure in Indian sheep ascertained using microsatellite information. Anim. Genet. 42:242–250. - PubMed
-
- Blackburn, H. D. , Toishibekov Y., Toishibekov M., Welsh C. S., Spiller S. F., Brown M., et al. 2011a. Genetic diversity of Ovis aries populations near domestication centers and in the New World. Genetica 139:1169–1178. - PubMed
-
- Blackburn, H. D. , Paiva S. R., Wildeus S., Getz W., Waldron D., Stobart R., et al. 2011b. Genetic structure and diversity among sheep breeds in the United States: identification of the major gene pools. J. Anim. Sci. 89:2336–2348. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

