Inferring Growth Control Mechanisms in Growing Multi-cellular Spheroids of NSCLC Cells from Spatial-Temporal Image Data
- PMID: 26866479
- PMCID: PMC4750943
- DOI: 10.1371/journal.pcbi.1004412
Inferring Growth Control Mechanisms in Growing Multi-cellular Spheroids of NSCLC Cells from Spatial-Temporal Image Data
Abstract
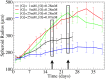
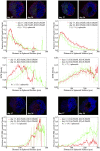
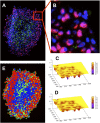
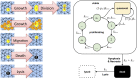
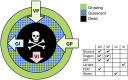
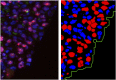
We develop a quantitative single cell-based mathematical model for multi-cellular tumor spheroids (MCTS) of SK-MES-1 cells, a non-small cell lung cancer (NSCLC) cell line, growing under various nutrient conditions: we confront the simulations performed with this model with data on the growth kinetics and spatial labeling patterns for cell proliferation, extracellular matrix (ECM), cell distribution and cell death. We start with a simple model capturing part of the experimental observations. We then show, by performing a sensitivity analysis at each development stage of the model that its complexity needs to be stepwise increased to account for further experimental growth conditions. We thus ultimately arrive at a model that mimics the MCTS growth under multiple conditions to a great extent. Interestingly, the final model, is a minimal model capable of explaining all data simultaneously in the sense, that the number of mechanisms it contains is sufficient to explain the data and missing out any of its mechanisms did not permit fit between all data and the model within physiological parameter ranges. Nevertheless, compared to earlier models it is quite complex i.e., it includes a wide range of mechanisms discussed in biological literature. In this model, the cells lacking oxygen switch from aerobe to anaerobe glycolysis and produce lactate. Too high concentrations of lactate or too low concentrations of ATP promote cell death. Only if the extracellular matrix density overcomes a certain threshold, cells are able to enter the cell cycle. Dying cells produce a diffusive growth inhibitor. Missing out the spatial information would not permit to infer the mechanisms at work. Our findings suggest that this iterative data integration together with intermediate model sensitivity analysis at each model development stage, provide a promising strategy to infer predictive yet minimal (in the above sense) quantitative models of tumor growth, as prospectively of other tissue organization processes. Importantly, calibrating the model with two nutriment-rich growth conditions, the outcome for two nutriment-poor growth conditions could be predicted. As the final model is however quite complex, incorporating many mechanisms, space, time, and stochastic processes, parameter identification is a challenge. This calls for more efficient strategies of imaging and image analysis, as well as of parameter identification in stochastic agent-based simulations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array [Journal Article]. Analyst. 2011;136(3):473–8. Available from: http://pubs.rsc.org/en/Content/ArticleLanding/2011/AN/ c0an00609b. 10.1039/C0AN00609B - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

