The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research
- PMID: 26866580
- PMCID: PMC4981567
- DOI: 10.1038/gim.2015.187
The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research
Abstract
Purpose: Geisinger Health System (GHS) provides an ideal platform for Precision Medicine. Key elements are the integrated health system, stable patient population, and electronic health record (EHR) infrastructure. In 2007, Geisinger launched MyCode, a system-wide biobanking program to link samples and EHR data for broad research use.
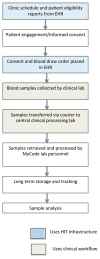
Methods: Patient-centered input into MyCode was obtained using participant focus groups. Participation in MyCode is based on opt-in informed consent and allows recontact, which facilitates collection of data not in the EHR and, since 2013, the return of clinically actionable results to participants. MyCode leverages Geisinger's technology and clinical infrastructure for participant tracking and sample collection.
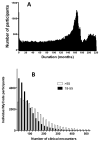
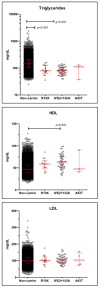
Results: MyCode has a consent rate of >85%, with more than 90,000 participants currently and with ongoing enrollment of ~4,000 per month. MyCode samples have been used to generate molecular data, including high-density genotype and exome sequence data. Genotype and EHR-derived phenotype data replicate previously reported genetic associations.
Conclusion: The MyCode project has created resources that enable a new model for translational research that is faster, more flexible, and more cost-effective than traditional clinical research approaches. The new model is scalable and will increase in value as these resources grow and are adopted across multiple research platforms.Genet Med 18 9, 906-913.
Figures
References
-
- Paulus RA, David K, Steele GD. Continuous innovation in health care: Implications of the geisinger experience. Health Aff (Millwood) 2008;27(5):1235–1245. - PubMed
-
- Casale AS, Paulua RA, Selna MJ, et al. “ProvenCareSM” A provider-driven pay-for-performance program for acute episodic cardiac surgical care. Annals of Surgery. 2007;249:613–623. - PubMed
-
- Precision medicine initiative. www.nih.gov/precisionmedicine. Updated 2015.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

