Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2
- PMID: 26866790
- PMCID: PMC4863649
- DOI: 10.1038/nprot.2016.018
Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2
Abstract
This protocol describes a method to obtain spatially resolved proteomic maps of specific compartments within living mammalian cells. An engineered peroxidase, APEX2, is genetically targeted to a cellular region of interest. Upon the addition of hydrogen peroxide for 1 min to cells preloaded with a biotin-phenol substrate, APEX2 generates biotin-phenoxyl radicals that covalently tag proximal endogenous proteins. Cells are then lysed, and biotinylated proteins are enriched with streptavidin beads and identified by mass spectrometry. We describe the generation of an appropriate APEX2 fusion construct, proteomic sample preparation, and mass spectrometric data acquisition and analysis. A two-state stable isotope labeling by amino acids in cell culture (SILAC) protocol is used for proteomic mapping of membrane-enclosed cellular compartments from which APEX2-generated biotin-phenoxyl radicals cannot escape. For mapping of open cellular regions, we instead use a 'ratiometric' three-state SILAC protocol for high spatial specificity. Isotopic labeling of proteins takes 5-7 cell doublings. Generation of the biotinylated proteomic sample takes 1 d, acquiring the mass spectrometric data takes 2-5 d and analysis of the data to obtain the final proteomic list takes 1 week.
Figures

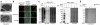
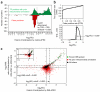
References
-
- Brunner Y, Schvartz D, Couté Y, Sanchez J-C. Proteomics of regulated secretory organelles. Mass Spectrom. Rev. 2009;28:844–867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

