Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism
- PMID: 26867183
- PMCID: PMC5271177
- DOI: 10.1016/j.chom.2016.01.002
Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism
Abstract
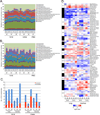
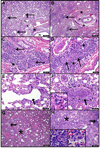
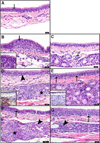
Pandemic influenza viruses modulate proinflammatory responses that can lead to immunopathogenesis. We present an extensive and systematic profiling of lipids, metabolites, and proteins in respiratory compartments of ferrets infected with either 1918 or 2009 human pandemic H1N1 influenza viruses. Integrative analysis of high-throughput omics data with virologic and histopathologic data uncovered relationships between host responses and phenotypic outcomes of viral infection. Proinflammatory lipid precursors in the trachea following 1918 infection correlated with severe tracheal lesions. Using an algorithm to infer cell quantity changes from gene expression data, we found enrichment of distinct T cell subpopulations in the trachea. There was also a predicted increase in inflammatory monocytes in the lung of 1918 virus-infected animals that was sustained throughout infection. This study presents a unique resource to the influenza research community and demonstrates the utility of an integrative systems approach for characterization of lipid metabolism alterations underlying respiratory responses to viruses.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




References
-
- Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, Wong G, Kobinger G, Xing Z, Couture C, et al. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity. 2014;40:554–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103493/GM/NIGMS NIH HHS/United States
- P51OD010425/OD/NIH HHS/United States
- U19 AI109761/AI/NIAID NIH HHS/United States
- P51 OD010425/OD/NIH HHS/United States
- HHSN272201400005C/PHS HHS/United States
- HHSN272201400006C/AI/NIAID NIH HHS/United States
- HHSN272200800060C/AI/NIAID NIH HHS/United States
- U19AI109761/AI/NIAID NIH HHS/United States
- T32 OD010423/OD/NIH HHS/United States
- HHSN272201400005C/AI/NIAID NIH HHS/United States
- 5T32OD010423-07/OD/NIH HHS/United States
- HHSN272201400006C/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

