Tripartite assembly of RND multidrug efflux pumps
- PMID: 26867482
- PMCID: PMC4754349
- DOI: 10.1038/ncomms10731
Tripartite assembly of RND multidrug efflux pumps
Abstract
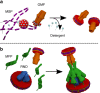
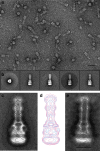
Tripartite multidrug efflux systems of Gram-negative bacteria are composed of an inner membrane transporter, an outer membrane channel and a periplasmic adaptor protein. They are assumed to form ducts inside the periplasm facilitating drug exit across the outer membrane. Here we present the reconstitution of native Pseudomonas aeruginosa MexAB-OprM and Escherichia coli AcrAB-TolC tripartite Resistance Nodulation and cell Division (RND) efflux systems in a lipid nanodisc system. Single-particle analysis by electron microscopy reveals the inner and outer membrane protein components linked together via the periplasmic adaptor protein. This intrinsic ability of the native components to self-assemble also leads to the formation of a stable interspecies AcrA-MexB-TolC complex suggesting a common mechanism of tripartite assembly. Projection structures of all three complexes emphasize the role of the periplasmic adaptor protein as part of the exit duct with no physical interaction between the inner and outer membrane components.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

