The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis
- PMID: 26867493
- PMCID: PMC4874886
- DOI: 10.1016/j.jhep.2016.01.035
The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis
Abstract
Background & aims: Alcoholic liver disease (ALD) ranges from fatty liver to inflammation and cirrhosis. miRNA-155 is an important regulator of inflammation. In this study, we describe the in vivo role of miR-155 in ALD.
Methods: Wild-type (WT) (C57/BL6J) or miR-155 knockout (KO) and TLR4 KO mice received Lieber DeCarli diet for 5weeks. Some mice received corn oil or CCl4 for 2 or 9weeks.
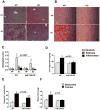
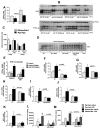
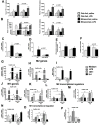
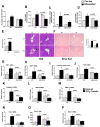
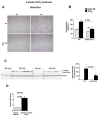
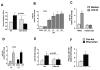
Results: We found that miR-155 KO mice are protected from alcohol-induced steatosis and inflammation. The reduction in alcohol-induced fat accumulation in miR-155 KO mice was associated with increased peroxisome proliferator-activated receptor response element (PPRE) and peroxisome proliferator-activated receptors (PPAR)α (miR-155 target) binding and decreased MCP1 production. Treatment with a miR-155 inhibitor increased PPARγ expression in naïve and alcohol treated RAW macrophages. Alcohol increased lipid metabolism gene expression (FABP4, LXRα, ACC1 and LDLR) in WT mice and this was prevented in KO mice. Alcohol diet caused an increase in the number of CD163(+) CD206(+) infiltrating macrophages and neutrophils in WT mice, which was prevented in miR-155 KO mice. Kupffer cells isolated from miR-155 KO mice exhibited predominance of M2 phenotype when exposed to M1 polarized signals and this was due to increased C/EBPβ. Pro-fibrotic genes were attenuated in miR-155 KO mice after alcohol diet or CCl4 treatment. Compared to WT mice, attenuation in CCl4 induced hydroxyproline and α-SMA was observed in KO mice. Finally, we show TLR4 signaling regulates miR-155 as TLR4 KO mice showed no induction of miR-155 after alcohol diet.
Conclusions: Collectively our results demonstrated the role of miR-155 in alcohol-induced steatohepatitis and fibrosis in vivo.
Keywords: Alcohol; Fibrosis; Inflammation; PPARα; PPARγ; microRNA.
Copyright © 2016 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest: None
Figures
Comment in
-
Tiny RNA with great effects: miR-155 in alcoholic liver disease.J Hepatol. 2016 Jun;64(6):1214-6. doi: 10.1016/j.jhep.2016.02.039. Epub 2016 Mar 3. J Hepatol. 2016. PMID: 26948494 No abstract available.
Similar articles
-
MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis.PLoS One. 2015 Jun 4;10(6):e0129251. doi: 10.1371/journal.pone.0129251. eCollection 2015. PLoS One. 2015. PMID: 26042593 Free PMC article.
-
The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88.Hepatology. 2008 Oct;48(4):1224-31. doi: 10.1002/hep.22470. Hepatology. 2008. PMID: 18792393 Free PMC article.
-
Chronic alcohol-induced microRNA-155 contributes to neuroinflammation in a TLR4-dependent manner in mice.PLoS One. 2013 Aug 9;8(8):e70945. doi: 10.1371/journal.pone.0070945. eCollection 2013. PLoS One. 2013. PMID: 23951048 Free PMC article.
-
Roles of peroxisome proliferator-activated receptor α in the pathogenesis of ethanol-induced liver disease.Chem Biol Interact. 2020 Aug 25;327:109176. doi: 10.1016/j.cbi.2020.109176. Epub 2020 Jun 11. Chem Biol Interact. 2020. PMID: 32534989 Review.
-
IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice.J Clin Invest. 2012 Oct;122(10):3476-89. doi: 10.1172/JCI60777. Epub 2012 Sep 4. J Clin Invest. 2012. PMID: 22945633 Free PMC article. Review.
Cited by
-
Effect of procyanidins on lipid metabolism and inflammation in rats exposed to alcohol and iron.Heliyon. 2020 Sep 7;6(9):e04847. doi: 10.1016/j.heliyon.2020.e04847. eCollection 2020 Sep. Heliyon. 2020. PMID: 32964156 Free PMC article.
-
Role of cuproptosis-related gene in lung adenocarcinoma.Front Oncol. 2022 Dec 21;12:1080985. doi: 10.3389/fonc.2022.1080985. eCollection 2022. Front Oncol. 2022. PMID: 36620594 Free PMC article.
-
Mulberry fruit repairs alcoholic liver injury by modulating lipid metabolism and the expression of miR-155 and PPARα in rats.Funct Integr Genomics. 2023 Aug 2;23(3):261. doi: 10.1007/s10142-023-01131-y. Funct Integr Genomics. 2023. PMID: 37530875
-
Dexamethasone relieves the inflammatory response caused by inguinal hernia meshes through miR-155.Hernia. 2024 Aug;28(4):1113-1119. doi: 10.1007/s10029-024-02985-2. Epub 2024 Mar 16. Hernia. 2024. PMID: 38492053
-
Detecting serum and urine metabolic profile changes of CCl4-liver fibrosis in rats at 12 weeks based on gas chromatography-mass spectrometry.Exp Ther Med. 2017 Aug;14(2):1496-1504. doi: 10.3892/etm.2017.4668. Epub 2017 Jun 26. Exp Ther Med. 2017. PMID: 28810615 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

