The Platelet Integrin αIIbβ3 Differentially Interacts with Fibrin Versus Fibrinogen
- PMID: 26867579
- PMCID: PMC4824994
- DOI: 10.1074/jbc.M115.706861
The Platelet Integrin αIIbβ3 Differentially Interacts with Fibrin Versus Fibrinogen
Abstract
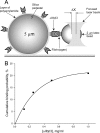
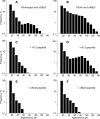
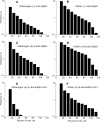
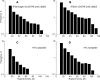
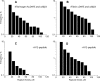
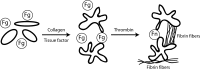
Fibrinogen binding to the integrin αIIbβ3 mediates platelet aggregation and spreading on fibrinogen-coated surfaces. However,in vivoαIIbβ3 activation and fibrinogen conversion to fibrin occur simultaneously, although the relative contributions of fibrinogenversusfibrin to αIIbβ3-mediated platelet functions are unknown. Here, we compared the interaction of αIIbβ3 with fibrin and fibrinogen to explore their differential effects. A microscopic bead coated with fibrinogen or monomeric fibrin produced by treating the immobilized fibrinogen with thrombin was captured by a laser beam and repeatedly brought into contact with surface-attached purified αIIbβ3. When αIIbβ3-ligand complexes were detected, the rupture forces were measured and displayed as force histograms. Monomeric fibrin displayed a higher probability of interacting with αIIbβ3 and a greater binding strength. αIIbβ3-fibrin interactions were also less sensitive to inhibition by abciximab and eptifibatide. Both fibrinogen- and fibrin-αIIbβ3 interactions were partially inhibited by RGD peptides, suggesting the existence of common RGD-containing binding motifs. This assumption was supported using the fibrin variants αD97E or αD574E with mutated RGD motifs. Fibrin made from a fibrinogen γ'/γ' variant lacking the γC αIIbβ3-binding motif was more reactive with αIIbβ3 than the parent fibrinogen. These results demonstrate that fibrin is more reactive with αIIbβ3 than fibrinogen. Fibrin is also less sensitive to αIIbβ3 inhibitors, suggesting that fibrin and fibrinogen have distinct binding requirements. In particular, the maintenance of αIIbβ3 binding activity in the absence of the γC-dodecapeptide and the α-chain RGD sequences suggests that the αIIbβ3-binding sites in fibrin are not confined to its known γ-chain and RGD motifs.
Keywords: adhesion; fibrin; fibrinogen; integrin; platelet.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Bennett J. S. (2001) Platelet-fibrinogen interactions. Ann. N.Y. Acad. Sci. 936, 340–354 - PubMed
-
- Collet J. P., Montalescot G., Lesty C., and Weisel J. W. (2002) A structural and dynamic investigation of the facilitating effect of glycoprotein IIb/IIIa inhibitors in dissolving platelet-rich clots. Circ. Res. 90, 428–434 - PubMed
-
- Carr M. E., Jr. (2003) Development of platelet contractile force as a research and clinical measure of platelet function. Cell Biochem. Biophys. 38, 55–78 - PubMed
-
- Hantgan R. R., and Mousa S. A. (1998) Inhibition of platelet-mediated clot retraction by integrin antagonists. Thromb. Res. 89, 271–279 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

