A variant at 9p21.3 functionally implicates CDKN2B in paediatric B-cell precursor acute lymphoblastic leukaemia aetiology
- PMID: 26868379
- PMCID: PMC4754340
- DOI: 10.1038/ncomms10635
A variant at 9p21.3 functionally implicates CDKN2B in paediatric B-cell precursor acute lymphoblastic leukaemia aetiology
Abstract
Paediatric B-cell precursor acute lymphoblastic leukaemia (BCP-ALL) is the most common cancer of childhood, yet little is known about BCP-ALL predisposition. In this study, in 2,187 cases of European ancestry and 5,543 controls, we discover and replicate a locus indexed by rs77728904 at 9p21.3 associated with BCP-ALL susceptibility (Pcombined=3.32 × 10(-15), OR=1.72) and independent from rs3731217, the previously reported ALL-associated variant in this region. Of correlated SNPs tagged by this locus, only rs662463 is significant in African Americans, suggesting it is a plausible causative variant. Functional analysis shows that rs662463 is a cis-eQTL for CDKN2B, with the risk allele associated with lower expression, and suggests that rs662463 influences BCP-ALL risk by regulating CDKN2B expression through CEBPB signalling. Functional analysis of rs3731217 suggests it is associated with BCP-ALL by acting within a splicing regulatory element determining CDKN2A exon 3 usage (P=0.01). These findings provide new insights into the critical role of the CDKN2 locus in BCP-ALL aetiology.
Figures
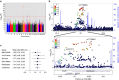

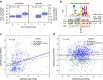

References
-
- Enciso-Mora V. et al.. Common genetic variation contributes significantly to the risk of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia 26, 2212–2215 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000430/TR/NCATS NIH HHS/United States
- CA40046/CA/NCI NIH HHS/United States
- P01 CA040046/CA/NCI NIH HHS/United States
- HD0433871/HD/NICHD NIH HHS/United States
- R21 CA129045/CA/NCI NIH HHS/United States
- U24 CA114766/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U01GM92666/GM/NIGMS NIH HHS/United States
- U10 CA098543/CA/NCI NIH HHS/United States
- R01 MH101820/MH/NIMH NIH HHS/United States
- CA129045/CA/NCI NIH HHS/United States
- U01CA176063/CA/NCI NIH HHS/United States
- U10 CA98543/CA/NCI NIH HHS/United States
- U01 GM092666/GM/NIGMS NIH HHS/United States
- P50 GM115279/GM/NIGMS NIH HHS/United States
- U01 CA176063/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

