Duplication and Diversification of Dipteran Argonaute Genes, and the Evolutionary Divergence of Piwi and Aubergine
- PMID: 26868596
- PMCID: PMC4824172
- DOI: 10.1093/gbe/evw018
Duplication and Diversification of Dipteran Argonaute Genes, and the Evolutionary Divergence of Piwi and Aubergine
Abstract
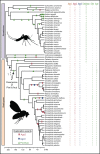
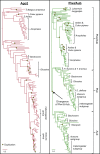
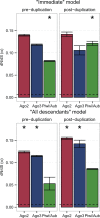
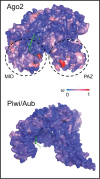
Genetic studies of Drosophila melanogaster have provided a paradigm for RNA interference (RNAi) in arthropods, in which the microRNA and antiviral pathways are each mediated by a single Argonaute (Ago1 and Ago2) and germline suppression of transposable elements is mediated by a trio of Piwi-subfamily Argonaute proteins (Ago3, Aub, and Piwi). Without a suitable evolutionary context, deviations from this can be interpreted as derived or idiosyncratic. Here we analyze the evolution of Argonaute genes across the genomes and transcriptomes of 86 Dipteran species, showing that variation in copy number can occur rapidly, and that there is constant flux in some RNAi mechanisms. The lability of the RNAi pathways is illustrated by the divergence of Aub and Piwi (182-156 Ma), independent origins of multiple Piwi-family genes in Aedes mosquitoes (less than 25Ma), and the recent duplications of Ago2 and Ago3 in the tsetse fly Glossina morsitans. In each case the tissue specificity of these genes has altered, suggesting functional divergence or innovation, and consistent with the action of dynamic selection pressures across the Argonaute gene family. We find there are large differences in evolutionary rates and gene turnover between pathways, and that paralogs of Ago2, Ago3, and Piwi/Aub show contrasting rates of evolution after duplication. This suggests that Argonautes undergo frequent evolutionary expansions that facilitate functional divergence.
Keywords: Argonaute; Diptera; Piwi; RNAi; gene duplication.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Anisimova M, Bielawski JP, Yang Z. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 18:1585–1592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

