Selective inhibition of EZH2 by ZLD1039 blocks H3K27 methylation and leads to potent anti-tumor activity in breast cancer
- PMID: 26868841
- PMCID: PMC4751454
- DOI: 10.1038/srep20864
Selective inhibition of EZH2 by ZLD1039 blocks H3K27 methylation and leads to potent anti-tumor activity in breast cancer
Abstract
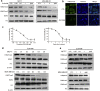
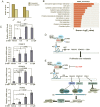
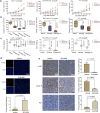
Enhancer of zeste homolog 2 (EZH2) is a candidate oncogenic driver due to its prevalent overexpression and aberrant repression of tumor suppressor genes in diverse cancers. Therefore, blocking EZH2 enzyme activity may present a valid therapeutic strategy for the treatment of cancers with EZH2 overexpression including breast cancers. Here, we described ZLD1039 a potent, highly selective, and orally bioavailable small molecule inhibitor of EZH2, which inhibited breast tumor growth and metastasis. ZLD1039 considerably inhibited EZH2 methyltransferase activity with nanomolar potency, decreased global histone-3 lysine-27 (H3K27) methylation, and reactivated silenced tumor suppressors connected to increased survival of patients with breast cancer. Comparable to conditional silencing of EZH2, its inhibition by ZLD1039 decreased cell proliferation, cell cycle arrest, and induced apoptosis. Comparably, treatment of xenograft-bearing mice with ZLD1039 led to tumor growth regression and metastasis inhibition. These data confirmed the dependency of breast cancer progression on EZH2 activity and the usefulness of ZLD1039 as a promising treatment for breast cancer.
Figures




Similar articles
-
Selective inhibition of EZH2 by ZLD10A blocks H3K27 methylation and kills mutant lymphoma cells proliferation.Biomed Pharmacother. 2016 Jul;81:288-294. doi: 10.1016/j.biopha.2016.04.019. Epub 2016 Apr 25. Biomed Pharmacother. 2016. PMID: 27261606
-
Selective EZH2 inhibitor zld1039 alleviates inflammation in cisplatin-induced acute kidney injury partially by enhancing RKIP and suppressing NF-κB p65 pathway.Acta Pharmacol Sin. 2022 Aug;43(8):2067-2080. doi: 10.1038/s41401-021-00837-8. Epub 2021 Dec 22. Acta Pharmacol Sin. 2022. PMID: 34937916 Free PMC article.
-
Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation.Proc Natl Acad Sci U S A. 2012 Dec 26;109(52):21360-5. doi: 10.1073/pnas.1210371110. Epub 2012 Dec 10. Proc Natl Acad Sci U S A. 2012. PMID: 23236167 Free PMC article.
-
EZH2: a novel target for cancer treatment.J Hematol Oncol. 2020 Jul 28;13(1):104. doi: 10.1186/s13045-020-00937-8. J Hematol Oncol. 2020. PMID: 32723346 Free PMC article. Review.
-
EZH2 in normal hematopoiesis and hematological malignancies.Oncotarget. 2016 Jan 19;7(3):2284-96. doi: 10.18632/oncotarget.6198. Oncotarget. 2016. PMID: 26497210 Free PMC article. Review.
Cited by
-
Dysregulation of histone methyltransferases in breast cancer - Opportunities for new targeted therapies?Mol Oncol. 2016 Dec;10(10):1497-1515. doi: 10.1016/j.molonc.2016.09.003. Epub 2016 Sep 23. Mol Oncol. 2016. PMID: 27717710 Free PMC article. Review.
-
CBX6 is negatively regulated by EZH2 and plays a potential tumor suppressor role in breast cancer.Sci Rep. 2019 Jan 17;9(1):197. doi: 10.1038/s41598-018-36560-4. Sci Rep. 2019. PMID: 30655550 Free PMC article.
-
Lysine methyltransferase inhibitors: where we are now.RSC Chem Biol. 2021 Dec 13;3(4):359-406. doi: 10.1039/d1cb00196e. eCollection 2022 Apr 6. RSC Chem Biol. 2021. PMID: 35441141 Free PMC article. Review.
-
Epigenetic modifiers in immunotherapy: a focus on checkpoint inhibitors.Immunotherapy. 2016 Jun;8(6):705-19. doi: 10.2217/imt-2016-0014. Immunotherapy. 2016. PMID: 27197539 Free PMC article. Review.
-
Targeting Histone Methyltransferase DOT1L by a Novel Psammaplin A Analog Inhibits Growth and Metastasis of Triple-Negative Breast Cancer.Mol Ther Oncolytics. 2019 Oct 1;15:140-152. doi: 10.1016/j.omto.2019.09.005. eCollection 2019 Dec 20. Mol Ther Oncolytics. 2019. PMID: 31720371 Free PMC article.
References
-
- Kouzarides T. Chromatin modifications and their function. Cell 128, 693–705 (2007). - PubMed
-
- Helin K. & Dhanak D. Chromatin proteins and modifications as drug targets. Nature 502, 480–488 (2013). - PubMed
-
- Kubicek S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

