Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon
- PMID: 26869021
- PMCID: PMC4751504
- DOI: 10.1038/srep21623
Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon
Abstract
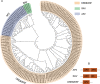
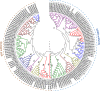
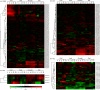
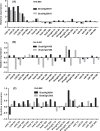
APETALA2/ethylene-responsive element binding protein (AP2/EREBP) transcription factors constitute one of the largest and most conserved gene families in plant, and play essential roles in growth, development and stress response. Except a few members, the AP2/EREBP family has not been characterized in Brachypodium distachyon, a model plant of Poaceae. We performed a genome-wide study of this family in B. distachyon by phylogenetic analyses, transactivation assays and transcript profiling. A total of 149 AP2/EREBP genes were identified and divided into four subfamilies, i.e., ERF (ethylene responsive factor), DREB (dehydration responsive element binding gene), RAV (related to ABI3/VP) and AP2. Tandem duplication was a major force in expanding B. distachyon AP2/EREBP (BdAP2/EREBP) family. Despite a significant expansion, genomic organizations of BdAP2/EREBPs were monotonous as the majority of them, except those of AP2 subfamily, had no intron. An analysis of transcription activities of several closely related and duplicated BdDREB genes showed their functional divergence and redundancy in evolution. The expression of BdAP2/EREBPs in different tissues and the expression of DREB/ERF subfamilies in B. distachyon, wheat and rice under abiotic stresses were investigated by next-generation sequencing and microarray profiling. Our results are valuable for further function analysis of stress tolerant AP2/EREBP genes in B. distachyon.
Figures



Similar articles
-
Expansion and stress responses of the AP2/EREBP superfamily in cotton.BMC Genomics. 2017 Jan 31;18(1):118. doi: 10.1186/s12864-017-3517-9. BMC Genomics. 2017. PMID: 28143399 Free PMC article.
-
Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in Brachypodium distachyon.BMC Genomics. 2016 Aug 15;17(1):636. doi: 10.1186/s12864-016-2968-8. BMC Genomics. 2016. PMID: 27527343 Free PMC article.
-
Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice.Plant Cell Physiol. 2011 Feb;52(2):344-60. doi: 10.1093/pcp/pcq196. Epub 2010 Dec 17. Plant Cell Physiol. 2011. PMID: 21169347
-
The AP2/EREBP family of plant transcription factors.Biol Chem. 1998 Jun;379(6):633-46. doi: 10.1515/bchm.1998.379.6.633. Biol Chem. 1998. PMID: 9687012 Review.
-
Advances in AP2/ERF super-family transcription factors in plant.Crit Rev Biotechnol. 2020 Sep;40(6):750-776. doi: 10.1080/07388551.2020.1768509. Epub 2020 Jun 10. Crit Rev Biotechnol. 2020. PMID: 32522044 Review.
Cited by
-
Reference genes for Eucalyptus spp. under Beauveria bassiana inoculation and subsequently infestation by the galling wasp Leptocybe invasa.Sci Rep. 2024 Jan 31;14(1):2556. doi: 10.1038/s41598-024-52948-x. Sci Rep. 2024. PMID: 38297150 Free PMC article.
-
Identification, Classification, and Functional Analysis of AP2/ERF Family Genes in the Desert Moss Bryum argenteum.Int J Mol Sci. 2018 Nov 19;19(11):3637. doi: 10.3390/ijms19113637. Int J Mol Sci. 2018. PMID: 30463185 Free PMC article.
-
Transcriptional and Metabolomic Analyses Indicate that Cell Wall Properties are Associated with Drought Tolerance in Brachypodium distachyon.Int J Mol Sci. 2019 Apr 10;20(7):1758. doi: 10.3390/ijms20071758. Int J Mol Sci. 2019. PMID: 30974727 Free PMC article.
-
SNP-based mixed model association of growth- and yield-related traits in popcorn.PLoS One. 2019 Jun 25;14(6):e0218552. doi: 10.1371/journal.pone.0218552. eCollection 2019. PLoS One. 2019. PMID: 31237892 Free PMC article.
-
Genome-Wide Identification of AP2/ERF Transcription Factors in Cauliflower and Expression Profiling of the ERF Family under Salt and Drought Stresses.Front Plant Sci. 2017 Jun 8;8:946. doi: 10.3389/fpls.2017.00946. eCollection 2017. Front Plant Sci. 2017. PMID: 28642765 Free PMC article.
References
-
- Dubouzet J. G. et al.. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33, 751–763 (2003). - PubMed
-
- Gutterson N. & Reuber T. L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7, 465–471 (2004). - PubMed
-
- Kume S. et al.. Differential and coordinated expression of Cbf and Cor/Lea genes during long-term cold acclimation in two wheat cultivars showing distinct levels of freezing tolerance. Genes Genet Syst 80, 185–197 (2005). - PubMed
-
- Yamaguchi-Shinozaki K. & Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57, 781–803 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

