ALS-linked protein disulfide isomerase variants cause motor dysfunction
- PMID: 26869642
- PMCID: PMC4972141
- DOI: 10.15252/embj.201592224
ALS-linked protein disulfide isomerase variants cause motor dysfunction
Abstract
Disturbance of endoplasmic reticulum (ER) proteostasis is a common feature of amyotrophic lateral sclerosis (ALS). Protein disulfide isomerases (PDIs) areERfoldases identified as possibleALSbiomarkers, as well as neuroprotective factors. However, no functional studies have addressed their impact on the disease process. Here, we functionally characterized fourALS-linked mutations recently identified in two majorPDIgenes,PDIA1 andPDIA3/ERp57. Phenotypic screening in zebrafish revealed that the expression of thesePDIvariants induce motor defects associated with a disruption of motoneuron connectivity. Similarly, the expression of mutantPDIs impaired dendritic outgrowth in motoneuron cell culture models. Cellular and biochemical studies identified distinct molecular defects underlying the pathogenicity of thesePDImutants. Finally, targetingERp57 in the nervous system led to severe motor dysfunction in mice associated with a loss of neuromuscular synapses. This study identifiesERproteostasis imbalance as a risk factor forALS, driving initial stages of the disease.
Keywords: ERp57; PDIA1; amyotrophic lateral sclerosis; protein disulfide isomerase.
© 2016 The Authors.
Figures

- A
Location of missense variants of
PDIA 1 andER p57 identified inALS cases.PDI primary structure: catalytic a and a′ domains containing the active‐site motifCXXC sequence (dark purple and dark green), non‐catalytic domains b and b′ containing ligand‐binding sites (light purple and light green), and x‐linker region (light gray). The constructs used in this study contained a V5‐tag (orange) at the C‐terminus that was inserted previous to theER ‐retention signal (dark gray). - B
Expression of
PDI variants in zebrafish. Zebrafish embryos at 1–2‐cell stage were injected with sensemRNA coding for the indicatedPDI s (PDIA 1WT ,PDIA 1D292N, andPDIA 1R300H: 200 pg/embryo;ER p57WT ,ER p57D217N, andER p57Q481K: 80 pg/embryo). Protein expression was confirmed by Western blot analysis using anti‐V5 antibody in total embryo extracts at 24 h post‐fertilization (hpf). Actin was used as a loading control. In the left panel, 80 μg protein extract was used, and in the right panel, 120 μg protein extract was used. - C, D
Motoneuron defects induced in zebrafish embryos after expression of the indicated
ALS ‐linkedPDI mutants and wild‐type controls (PDIA 1WT andPDIA 1R300H: 80 pgmRNA /embryo;ER p57WT ,ER p57D217N,ER p57D217N, andER p57Q481K: 30 pgmRNA /embryo). The most frequent global phenotypes induced byPDI injection are shown in lateral views of embryos at 24 hpf (left column). Black arrows indicate the presence of curly tail and/or shorter axis phenotypes (see details in Appendix Table S1). Axon motoneuron morphology was visualized using confocal microscopy in lateral views of the trunk region in transgenic Tg(Huc:Kaede) zebrafish at 36 hpf (middle and right columns). Images in the right column correspond to magnification views of the rectangular regions depicted in left and middle columns. Asterisks and arrows point to examples of increased axonal branching and reduced axonal length, respectively. - E, F
Quantification of motoneuron axon length and axon branching in 36 hpf Tg(Huc:Kaede) embryos injected with the indicated
PDIA 1 (E) andER p57 (F) mutants. - G
Touch‐evoked escape responses of 48 hpf zebrafish embryos injected with
ALS ‐linkedPDI mutants (Kabashi et al, 2009, 2011). The number of touches necessary to evoke an escape response (left) and speed (right, in mm/s) of the escape response was determined for each condition.


- A
NSC 34 cells were transiently transfected with expression vectors for V5‐tagged wild‐type and mutantPDI s. After 48 h, overexpressedPDI variants were assessed under reducing conditions in an 8%SDS –PAGE . An antibody detects totalPDIA 1 (endogenous mousePDIA 1 and exogenous human V5‐taggedPDIA 1) (first panel). A second antibody detects only humanPDIA 1, therefore only V5‐taggedPDIA 1 appears (second panel). A mouse‐ and human‐specific antibodies were used to detect totalER p57 (third panel). Anti‐V5 was used to detect the exogenousPDI variants. Anti‐β‐actin was used as a loading control. - B, C
NSC 34 cells were transiently transfected with the indicatedPDI constructs together with aGFP expression plasmid. Cells were then differentiated for 24 h in Neurobasal medium containing B27 supplement to induce cell differentiation. Increased neurite outgrowth is indicated with white arrow heads. (B) Quantification of the average primary neurite lengths was performed, all cells from three independent experiments were compiled. A minimum of 100 cells per experiment were analyzed. In addition, (C) the percentage of cells with neurites was quantified in the three independent experiments (right panel). - D
Primary rat ventral spinal cord neurons were prepared and after 4 days in vitro (
DIV ) transfected withGFP alone or together with the indicatedPDI constructs. Cells were fixed at 10DIV andSMI 32 staining was performed to identify motoneurons. Images were taken and the total outgrowth ofGFP ‐ andSMI 32‐positive cells was quantified. Results are compiled from three independent experiments. - E
Human motoneurons were differentiated from the human embryonic stem cell (
ESC ) HuESC 3 Hb9::GFP line. Differentiated neurons were transduced with lentivirus expressingGFP alone or together withPDI ‐expressing plasmids. Transduced cells were cultured for another 10 days. To identify motoneurons after lentiviral transduction, immunocytochemistry analyses were performed. Quantification of neurite outgrowth in the different experimental conditions ofGFP ‐ andHB 9‐positive neurons was obtained. Neurite number and assessment of the length of each neurite were performed in a similar manner as for primary rat motoneurons (see Materials and Methods). Four independent experiments were performed. - F
NSC 34 cell lines stably knocked down forPDIA 1 orER p57 were differentiated for 24 h in Neurobasal medium containing B27 supplement to induce cell differentiation. The percentage of cells with neurites was quantified in the three independent experiments. A minimum of 100 cells per experiment were analyzed.

- A
V5‐tagged wild‐type and mutant
PDI s were transiently expressed inNSC 34 cells. After 44 h, cells were treated with 1 μg/ml tunicamycin for 16 h and theER stress markersXBP 1s,ATF 4, and BiP were detected using the respective antibodies. V5‐taggedPDI s were detected in the cell extract using anti‐V5.HSP 90 was used as a loading control. - B, C
Cell viability after treatment with thapsigargin (B) or tunicamycin (1 μg/ml) (C) was determined by the
MTT assay. The dose–response curve of the mock is the same, since all treatments were performed in the same experiment. Mean andSEM of triplicate measurements of one representative experiment are shown. - D
BDNF ‐GFP was co‐expressed with V5‐taggedPDI s (wild types and mutants) in N2a cells.BDNF secretion to the cell medium was assessed by Western blot after 18 and 42 h. As a control, V5‐taggedPDI s were detected in the cell extract using anti‐V5. - E
V5‐tagged wild‐type and mutant
PDI s were overexpressed in Neuro2a cells. Cell medium was collected after 48 h and tested for the presence of progranulin byELISA .

PDI mutants form abnormal disulfide‐dependent protein complexes.NSC 34 cells were transiently transfected with expression vectors for V5‐tagged wild‐type and mutantPDIA 1. After 48 h, differential disulfide‐dependent interactions/aggregations of overexpressedPDI variants was assessed under reducing (+DTT ) and non‐reducing (−DTT ) conditions in an 8%SDS –PAGE . Anti‐V5 was used for detection in Western blot.Analysis of the
PDIA 1 structure to model the effects of the R300H mutation. The close association between Arg300 located to the b′ domain ofPDIA 1 with Trp396 located to the a′ domain adjacent to the active‐site motifCGHC (designated asAS in yellow) is shown in comparison with the mutated version ofPDIA 1R300H highlighting the same residues. A potential stabilization of the interaction between the b′ and a′ domain is shown that may be caused by the interaction of the imidazole rings of mutated His300 with Trp396.ALS ‐linkedPDIA 1 variants were generated as recombinant proteins and then analyzed by circular dichroism (CD ). Averages forCD spectroscopic scans ofPDIA 1WT and mutants are shown.Averages for
CD spectroscopic thermal denaturation of recombinantPDIA 1WT and mutants.Representative electrophoresis analysis of proteinase K‐treated recombinant
PDIA 1 variants. Mass spectrometric analysis of proteinase K‐digested samples from total sample and from in‐gel trypsin‐digested protein samples (arrows indicate band 1 and band 2) indicated differences in the removal of the x‐linker region. Bottom panel: ratios of band 1 to band 2 were quantified in four independent experiments.Representative fluorescence spectra of recombinant
PDIA 1 b′xWT fragments and the equivalentALS ‐linked mutants (n = 6). Ratios from the two peak areas and change compared toPDIA 1 b′xWT . Both mutants analyzed show a significant shift in peak position compared to wild type, but in opposite directions, suggesting thatPDIA 1D292N shifts equilibrium toward the capped version (x‐region over the binding pocket) and thePDIA 1R300H mutant toward the uncapped version (Nguyen et al, 2008).The activity of recombinant
PDIA 1 was measured in vitro using aBPTI refolding assay following by mass spectrometry analysis. The percentages of differentBPTI species was calculated (6H, fully reduced; 1S, one disulfide bond; 2S, two disulfide bonds; 3S, three disulfide bonds) at time point 2.5 min in four independent experiments.Measurement of H2O2 levels at the
ER lumen of living cells. Left panel: reduced PDIs can be oxidized by ERO1Lα, which then transfers electrons to molecular oxygen (O2) to generate hydrogen peroxide as product from PDI activity. Right panel:NSC 34 cells were transiently co‐transfected withER luminal HyPer sensor and indicatedPDI s. After 48 h, the 490/420 nm fluorescence ratio was recorded for 2 min under basal conditions. Means andSEM derived from all cells per condition (n = 55–74) monitored in four independent experiments are shown.HEK 293T cells were transfected with expression vectors for V5‐tagged wild‐type and mutantPDIA 1, as well as empty vector. After 48 h, V5‐tagged proteins were immunoprecipitated and eluted with V5 peptide. The interaction with endogenousERO 1Lα was analyzed by Western blot. The inputs and elutions are shown as control. Right panel: quantification of the degree of interaction is presented.

PDI mutants form abnormal disulfide‐dependent protein complexes.NSC 34 cells were transiently transfected with expression vectors for V5‐tagged wild‐type and mutantER p57. After 48 h, differential disulfide‐dependent interactions/aggregations of overexpressedPDI variants was assessed under reducing (+DTT ) and non‐reducing (−DTT ) conditions in an 8%SDS –PAGE . Anti‐V5 was used for detection in Western blot.ALS ‐linkedER p57 variants were generated as recombinant proteins and then analyzed by circular dichroism (CD ). Averages forCD spectroscopic scans of recombinantER p57WT and mutants are shown.A thermal denaturation curve of
ER p57WT and mutants was performed.Representative electrophoresis of proteinase K‐treated
ER p57 recombinant proteins.Surface plasmon resonance analysis was performed to monitor the affinity of recombinant
ER p57 for recombinantCRT P domain.KD values are expressed as percentage ofER p57WT . Values are derived from four independent experiments. For absoluteKD values, please see Appendix Table S2.HEK 293T cells were transfected with expression vectors for V5‐tagged wild‐type and mutantER p57, as well as empty vector. After 48 h, V5‐tagged proteins were immunoprecipitated and eluted with V5 peptide. The interaction with endogenous calnexin (CNX ) and calreticulin (CRT ) was analyzed by Western blot. The inputs and elutions are shown. Right panel: quantification of the degree of interaction is presented.The gain of an N‐glycosylation site of
ER p57D217N was predicted after the analysis of the protein sequence since the change of Asp217 to an Asn creates theNXT /S consensus sequence. Neuro2a cells were transfected with expression vectors for V5‐tagged wild‐type and mutantER p57, as well as empty vector and treated with 1 μg/ml tunicamycin (Tm) for 20 h to inhibit N‐glycosylation. Alternatively, protein extracts were digested withPNG ase F and the possible removal of the N‐glycosylation was analyzed by Western blot using anti‐V5 antibody.

ER p57flox/flox mice were crossed with Nestin‐Cre transgenic mice to generate nervous system‐specificER p57‐deficient animals. The levels ofER p57 protein in the spinal cord were monitored by Western blot.ER p57WT : n = 4,ER p57Nes+/−: n = 5, andER p57Nes−/−: n = 4 mice.HSP 90 levels were used as a loading control. Right panel: quantification ofER p57 levels was performed relative to Hsp90 levels.Body weight measurements were performed at the indicated time points in
ER p57WT (n = 50),ER p57Nes+/− (n = 32) andER p57Nes−/− (n = 19) mice.Rotarod performance was performed
ER p57WT (n = 20),ER p57Nes+/− (n = 15) andER p57Nes−/− (n = 8) mice.Hanging test performance was assessed in
ER p57WT (n = 41),ER p57Nes+/− (n = 32), andER p57Nes−/− (n = 12) mice.Kaplan–Meier survival curve for
ER p57Nes−/− mice (N = 19) that prematurely died or had to be sacrificed because of health reasons between the ages 22 and 73 days. Mean survival of this subgroup of animals was 57 days.ER p57WT (n = 50) andER p57Nes+/− (n = 32) mice are shown as a reference.Histological analysis of NeuN and
GFAP staining was performed in spinal cord tissue fromER p57WT andER p57Nes−/− mice in three animals per group using indirect immunofluorescence. The nucleus was stained with Hoechst. Representative images from one mouse per group are shown. Scale bar: 50 μm.Stereological analysis of the spinal cord from
ER p57WT (n = 4),ER p57Nes+/− (n = 4) andER p57Nes−/− (n = 4) mice. Alternate series of sections from the spinal cord of the mice were either stained for Nissl (top row images) or processed for immunohistochemistry for the cholinergic cell marker choline acetyltransferase (ChAT , bottom row images). The nucleoli of the motoneurons, as stained in the Nissl series, were counted inside the motoneurons pools previously defined using the adjacent ChAT series (contours not shown). Cell densities of the three genotypes are shown on the right plots. No significant differences were found between the genotypes. Scale bar represents 200 μm and 50 μm on large and inset images, respectively.Representative images of the spinal cord myelination of
ER p57WT andER p57Nes−/− mice.
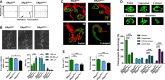
Representative
EMG recordings ofER p57WT (n = 16),ER p57Nes−/+ (n = 12) andER p57Nes−/− (n = 5) mice is presented. The presence of positive sharp waves (PSW ) inER p57Nes+/− andER p57Nes−/− mice indicated muscle denervation.Analysis of
NMJ and muscle morphologies were performed inER p57WT (n = 5),ER p57Nes+/− (n = 5) andER p57Nes−/− (n = 4) mice. Quantifications of endplate width, measuring the most ventral region of the diaphragms every 134 μm considering both sides of the innervation profile. Twenty to forty measurements per diaphragm were obtained per animal.Whole‐mounted diaphragms from
ER p57WT andER p57Nes−/− mice were co‐stained with anti‐neurofilament (red) and α‐BTX to reveal the postsynaptic densities (green). Three‐dimensional reconstructions (lower panel) of higher magnification are shown.ER p57WT NMJ s are fully innervated pretzel‐like, whereasER p57Nes−/−NMJ s display less complex postsynaptic densities and an incomplete or aberrantly distributed innervation pattern (Videos EV4, EV5, EV6, EV7).Representative 3D reconstructions of z‐stacks showing the five categories in which
NMJ morphologies were grouped: pretzel, fragmented, O‐shaped, C‐shaped, and dismantledNMJ s. Histogram of the distribution frequency of > 200 differentNMJ per animal is shown.Quantitative morphometry of > 60
NMJ s in the different genotypes analyzed.

The levels of
ER p57 protein in the muscle were monitored by Western blot.ER p57WT (n = 4),ER p57Nes+/− (n = 5), andER p57Nes−/− (n = 4) mice.HSP 90 was used as a loading control.Tibialis anterior muscles from
ER p57WT (n = 5),ER p57Nes+/− (n = 5), andER p57Nes−/− (n = 4) mice were dissected and cryosectioned (20 μm) for the analysis of several parameters of muscle physiology. Upper panel: hematoxylin/chromotrope (HA ) staining was performed to assess the gross morphology of the muscle. Middle panel: Bismarck brown staining to monitor skeletal muscles fibrosis. Lower panel: cryosections were then double stained with wheat germ agglutinin (WGA ) to visualize plasma membrane and nuclei. The absence of central nuclei in all genotypes suggests thatER p57 deficiency does not affect degeneration/regeneration cycles. Images are representative of each genotype and where acquired in the same anatomical region of theTA muscles.Tibialis anterior cryosections (20 μm) from
ER p57WT ,ER p57Nes+/−, andER p57Nes−/− mice were stained forNADH ‐TR activity to identify oxidative myofibers (left panel). Representative micrographs of transversal cryosections show variable proportions of light, intermediate and dark positive staining in the different genotypes:NADH ‐TR ‐positive (slow twitch) andNADH ‐TR ‐negative (fast twitch) myofibers were quantified as percentage of total for every genotype (upper right panel). The mean diameter of slow‐ and fast‐twitch muscle fibers was evaluated at the light microscope using the ImageJ software (lower right panel). Data represent the average ±SEM of > 300 fibers per animal.

The levels of synaptic proteins
SV 2,NMDAR 2A,PSD 95, and synaptophysin in the cortex were monitored by Western blot.ER p57WT (n = 4),ER p57Nes+/− (n = 5) andER p57Nes−/− (n = 4) mice.HSP 90 and β‐actin were used as loading controls.Quantification of
SV 2 monomer (left panel) andSV 2 high molecular weight species (right panel). β‐actin was used as the reference.
Comment in
-
ER strikes again: Proteostasis Dysfunction In ALS.EMBO J. 2016 Apr 15;35(8):798-800. doi: 10.15252/embj.201694117. Epub 2016 Mar 11. EMBO J. 2016. PMID: 26968985 Free PMC article.
References
-
- Andreu CI, Woehlbier U, Torres M, Hetz C (2012) Protein disulfide isomerases in neurodegeneration: from disease mechanisms to biomedical applications. FEBS Lett 586: 2826–2834 - PubMed
-
- Armstrong GA, Drapeau P (2013) Loss and gain of FUS function impair neuromuscular synaptic transmission in a genetic model of ALS. Hum Mol Genet 22: 4282–4292 - PubMed
-
- Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, Nunan J, Rembach A, Nagley P, Beart PM, Cheema SS, Horne MK (2006) Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein‐disulfide isomerase with superoxide dismutase 1. J Biol Chem 281: 30152–30165 - PubMed
-
- Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK (2008) Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis 30: 400–407 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

