Small Genetic Circuits and MicroRNAs: Big Players in Polymerase II Transcriptional Control in Plants
- PMID: 26869700
- PMCID: PMC4790873
- DOI: 10.1105/tpc.15.00852
Small Genetic Circuits and MicroRNAs: Big Players in Polymerase II Transcriptional Control in Plants
Abstract
RNA Polymerase II (Pol II) regulatory cascades involving transcription factors (TFs) and their targets orchestrate the genetic circuitry of every eukaryotic organism. In order to understand how these cascades function, they can be dissected into small genetic networks, each containing just a few Pol II transcribed genes, that generate specific signal-processing outcomes. Small RNA regulatory circuits involve direct regulation of a small RNA by a TF and/or direct regulation of a TF by a small RNA and have been shown to play unique roles in many organisms. Here, we will focus on small RNA regulatory circuits containing Pol II transcribed microRNAs (miRNAs). While the role of miRNA-containing regulatory circuits as modular building blocks for the function of complex networks has long been on the forefront of studies in the animal kingdom, plant studies are poised to take a lead role in this area because of their advantages in probing transcriptional and posttranscriptional control of Pol II genes. The relative simplicity of tissue- and cell-type organization, miRNA targeting, and genomic structure make the Arabidopsis thaliana plant model uniquely amenable for small RNA regulatory circuit studies in a multicellular organism. In this Review, we cover analysis, tools, and validation methods for probing the component interactions in miRNA-containing regulatory circuits. We then review the important roles that plant miRNAs are playing in these circuits and summarize methods for the identification of small genetic circuits that strongly influence plant function. We conclude by noting areas of opportunity where new plant studies are imminently needed.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures
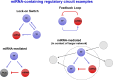

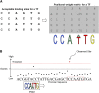
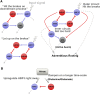

Comment in
-
The Plant Cell Reviews Small RNA and Chromatin Dynamics: From Small Genetic Circuits to Complex Genomes.Plant Cell. 2016 Feb;28(2):269-71. doi: 10.1105/tpc.16.00113. Epub 2016 Feb 11. Plant Cell. 2016. PMID: 26869698 Free PMC article. No abstract available.
References
-
- Achard P., Lagrange T., El-Zanaty A.-F., Mache R. (2003). Architecture and transcriptional activity of the initiator element of the TATA-less RPL21 gene. Plant J. 35: 743–752. - PubMed
-
- Alipanahi B., Delong A., Weirauch M.T., Frey B.J. (2015). Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33: 831–838. - PubMed
-
- Alon U. (2006). An Introduction to Systems Biology: Design Principles of Biological Circuits. (Boca Raton, FL: Chapman and Hall/CRC; ).
-
- Alon U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8: 450–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

