A Toolkit for Orthogonal and in vivo Optical Manipulation of Ionotropic Glutamate Receptors
- PMID: 26869877
- PMCID: PMC4735401
- DOI: 10.3389/fnmol.2016.00002
A Toolkit for Orthogonal and in vivo Optical Manipulation of Ionotropic Glutamate Receptors
Abstract
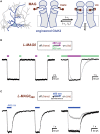
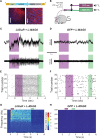
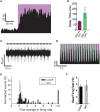
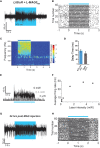
The ability to optically manipulate specific neuronal signaling proteins with genetic precision paves the way for the dissection of their roles in brain function, behavior, and disease. Chemical optogenetic control with photoswitchable tethered ligands (PTLs) enables rapid, reversible and reproducible activation or block of specific neurotransmitter-gated receptors and ion channels in specific cells. In this study, we further engineered and characterized the light-activated GluK2 kainate receptor, LiGluR, to develop a toolbox of LiGluR variants. Low-affinity LiGluRs allow for efficient optical control of GluK2 while removing activation by native glutamate, whereas variant RNA edited versions enable the synaptic role of receptors with high and low Ca(2+) permeability to be assessed and spectral variant photoswitches provide flexibility in illumination. Importantly, we establish that LiGluR works efficiently in the cortex of awake, adult mice using standard optogenetic techniques, thus opening the door to probing the role of specific synaptic receptors and cellular signals in the neural circuit operations of the mammalian brain in normal conditions and in disease. The principals developed in this study are widely relevant to the engineering and in vivo use of optically controllable proteins, including other neurotransmitter receptors.
Keywords: chemical optogenetics; glutamate receptor; in vivo; molecular engineering; photo-pharmacology.
Figures


Similar articles
-
Two-photon brightness of azobenzene photoswitches designed for glutamate receptor optogenetics.Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):E776-85. doi: 10.1073/pnas.1416942112. Epub 2015 Feb 4. Proc Natl Acad Sci U S A. 2015. PMID: 25653339 Free PMC article.
-
Probing the ionotropic activity of glutamate GluD2 receptor in HEK cells with genetically-engineered photopharmacology.Elife. 2020 Oct 28;9:e59026. doi: 10.7554/eLife.59026. Elife. 2020. PMID: 33112237 Free PMC article.
-
Optical control of ligand-gated ion channels.Methods Mol Biol. 2013;998:417-35. doi: 10.1007/978-1-62703-351-0_32. Methods Mol Biol. 2013. PMID: 23529448
-
Optogenetic pharmacology for control of native neuronal signaling proteins.Nat Neurosci. 2013 Jul;16(7):816-23. doi: 10.1038/nn.3424. Epub 2013 Jun 25. Nat Neurosci. 2013. PMID: 23799474 Free PMC article. Review.
-
[Optically dissecting brain nicotinic receptor function with photo-controllable designer receptors].Biol Aujourdhui. 2017;211(2):173-188. doi: 10.1051/jbio/2017022. Epub 2017 Dec 13. Biol Aujourdhui. 2017. PMID: 29236669 Review. French.
Cited by
-
Principles and Design of Molecular Tools for Sensing and Perturbing Cell Surface Receptor Activity.Chem Rev. 2025 Mar 12;125(5):2665-2702. doi: 10.1021/acs.chemrev.4c00582. Epub 2025 Feb 25. Chem Rev. 2025. PMID: 39999110 Review.
-
Optopharmacology reveals a differential contribution of native GABAA receptors to dendritic and somatic inhibition using azogabazine.Neuropharmacology. 2020 Oct 1;176:108135. doi: 10.1016/j.neuropharm.2020.108135. Epub 2020 May 21. Neuropharmacology. 2020. PMID: 32445639 Free PMC article.
-
A fine-tuned azobenzene for enhanced photopharmacology in vivo.Cell Chem Biol. 2021 Nov 18;28(11):1648-1663.e16. doi: 10.1016/j.chembiol.2021.02.020. Epub 2021 Mar 17. Cell Chem Biol. 2021. PMID: 33735619 Free PMC article.
-
Solid-Phase-Supported Chemoenzymatic Synthesis of a Light-Activatable tRNA Derivative.Angew Chem Int Ed Engl. 2022 Jan 3;61(1):e202111613. doi: 10.1002/anie.202111613. Epub 2021 Nov 22. Angew Chem Int Ed Engl. 2022. PMID: 34738704 Free PMC article.
-
Structure, Function, and Pharmacology of Glutamate Receptor Ion Channels.Pharmacol Rev. 2021 Oct;73(4):298-487. doi: 10.1124/pharmrev.120.000131. Pharmacol Rev. 2021. PMID: 34753794 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

