Applications of Bayesian Phylodynamic Methods in a Recent U.S. Porcine Reproductive and Respiratory Syndrome Virus Outbreak
- PMID: 26870024
- PMCID: PMC4735353
- DOI: 10.3389/fmicb.2016.00067
Applications of Bayesian Phylodynamic Methods in a Recent U.S. Porcine Reproductive and Respiratory Syndrome Virus Outbreak
Abstract
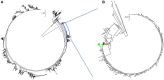
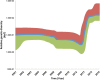
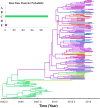
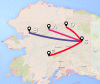
Classical phylogenetic methods such as neighbor-joining or maximum likelihood trees, provide limited inferences about the evolution of important pathogens and ignore important evolutionary parameters and uncertainties, which in turn limits decision making related to surveillance, control, and prevention resources. Bayesian phylodynamic models have recently been used to test research hypotheses related to evolution of infectious agents. However, few studies have attempted to model the evolutionary dynamics of porcine reproductive and respiratory syndrome virus (PRRSV) and, to the authors' knowledge, no attempt has been made to use large volumes of routinely collected data, sometimes referred to as big data, in the context of animal disease surveillance. The objective of this study was to explore and discuss the applications of Bayesian phylodynamic methods for modeling the evolution and spread of a notable 1-7-4 RFLP-type PRRSV between 2014 and 2015. A convenience sample of 288 ORF5 sequences was collected from 5 swine production systems in the United States between September 2003 and March 2015. Using coalescence and discrete trait phylodynamic models, we were able to infer population growth and demographic history of the virus, identified the most likely ancestral system (root state posterior probability = 0.95) and revealed significant dispersal routes (Bayes factor > 6) of viral exchange among systems. Results indicate that currently circulating viruses are evolving rapidly, and show a higher level of relative genetic diversity over time, when compared to earlier relatives. Biological soundness of model results is supported by the finding that sow farms were responsible for PRRSV spread within the systems. Such results cannot be obtained by traditional phylogenetic methods, and therefore, our results provide a methodological framework for molecular epidemiological modeling of new PRRSV outbreaks and demonstrate the prospects of phylodynamic models to inform decision-making processes for routine surveillance and, ultimately, to support prevention and control of food animal disease at local and regional scales.
Keywords: Bayesian phylodynamics; ORF5 gene; PRRSV; RFLP type 1-7-4; molecular surveillance.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

