RNA-SEQ Reveals Transcriptional Level Changes of Poplar Roots in Different Forms of Nitrogen Treatments
- PMID: 26870068
- PMCID: PMC4735414
- DOI: 10.3389/fpls.2016.00051
RNA-SEQ Reveals Transcriptional Level Changes of Poplar Roots in Different Forms of Nitrogen Treatments
Abstract
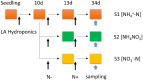
Poplar has emerged as a model plant for better understanding cellular and molecular changes accompanying tree growth, development, and response to environment. Long-term application of different forms of nitrogen (such as [Formula: see text]-N and [Formula: see text]-N) may cause morphological changes of poplar roots; however, the molecular level changes are still not well-known. In this study, we analyzed the expression profiling of poplar roots treated by three forms of nitrogen: S1 ([Formula: see text]), S2 (NH4NO3), and S3 ([Formula: see text]) by using RNA-SEQ technique. We found 463 genes significantly differentially expressed in roots by different N treatments, of which a total of 112 genes were found to differentially express between S1 and S2, 171 genes between S2 and S3, and 319 genes between S1 and S3. A cluster analysis shows significant difference in many transcription factor families and functional genes family under different N forms. Through an analysis of Mapman metabolic pathway, we found that the significantly differentially expressed genes are associated with fermentation, glycolysis, and tricarboxylic acid cycle (TCA), secondary metabolism, hormone metabolism, and transport processing. Interestingly, we did not find significantly differentially expressed genes in N metabolism pathway, mitochondrial electron transport/ATP synthesis and mineral nutrition. We also found abundant candidate genes (20 transcription factors and 30 functional genes) regulating morphology changes of poplar roots under the three N forms. The results obtained are beneficial to a better understanding of the potential molecular and cellular mechanisms regulating root morphology changes under different N treatments.
Keywords: RNA-SEQ; long-time nitrogen treatment; nitrogen forms; nitrogen metabolism; poplar.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

