BdCESA7, BdCESA8, and BdPMT Utility Promoter Constructs for Targeted Expression to Secondary Cell-Wall-Forming Cells of Grasses
- PMID: 26870070
- PMCID: PMC4740387
- DOI: 10.3389/fpls.2016.00055
BdCESA7, BdCESA8, and BdPMT Utility Promoter Constructs for Targeted Expression to Secondary Cell-Wall-Forming Cells of Grasses
Abstract
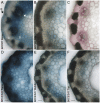
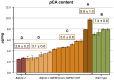
Utility vectors with promoters that confer desired spatial and temporal expression patterns are useful tools for studying gene and cellular function and for industrial applications. To target the expression of DNA sequences of interest to cells forming plant secondary cell walls, which generate most of the vegetative biomass, upstream regulatory sequences of the Brachypodium distachyon lignin biosynthetic gene BdPMT and the cellulose synthase genes BdCESA7 and BdCESA8 were isolated and cloned into binary vectors designed for Agrobacterium-mediated transformation of monocots. Expression patterns were assessed using the β-glucuronidase gene GUSPlus and X-glucuronide staining. All three promoters showed strong expression levels in stem tissue at the base of internodes where cell wall deposition is most active, in both vascular bundle xylem vessels and tracheids, and in interfascicular tissues, with expression less pronounced in developmentally older tissues. In leaves, BdCESA7 and BdCESA8 promoter-driven expression was strongest in leaf veins, leaf margins, and trichomes; relatively weaker and patchy expression was observed in the epidermis. BdPMT promoter-driven expression was similar to the BdCESA promoters expression patterns, including strong expression in trichomes. The intensity and extent of GUS staining varied considerably between transgenic lines, suggesting that positional effects influenced promoter activity. Introducing the BdPMT and BdCESA8 Open Reading Frames into BdPMT and BdCESA8 utility promoter binary vectors, respectively, and transforming those constructs into Brachypodium pmt and cesa8 loss-of-function mutants resulted in rescue of the corresponding mutant phenotypes. This work therefore validates the functionality of these utility promoter binary vectors for use in Brachypodium and likely other grass species. The identification, in Bdcesa8-1 T-DNA mutant stems, of an 80% reduction in crystalline cellulose levels confirms that the BdCESA8 gene is a secondary-cell-wall-forming cellulose synthase.
Keywords: Brachypodium; binary vectors; cellulose; lignin; monocot; p-coumarate; tissue-specific expression.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources

