Identification of fibrillogenic regions in human triosephosphate isomerase
- PMID: 26870617
- PMCID: PMC4748702
- DOI: 10.7717/peerj.1676
Identification of fibrillogenic regions in human triosephosphate isomerase
Abstract
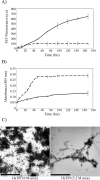
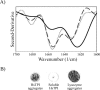
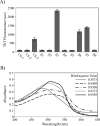
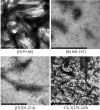
Background. Amyloid secondary structure relies on the intermolecular assembly of polypeptide chains through main-chain interaction. According to this, all proteins have the potential to form amyloid structure, nevertheless, in nature only few proteins aggregate into toxic or functional amyloids. Structural characteristics differ greatly among amyloid proteins reported, so it has been difficult to link the fibrillogenic propensity with structural topology. However, there are ubiquitous topologies not represented in the amyloidome that could be considered as amyloid-resistant attributable to structural features, such is the case of TIM barrel topology. Methods. This work was aimed to study the fibrillogenic propensity of human triosephosphate isomerase (HsTPI) as a model of TIM barrels. In order to do so, aggregation of HsTPI was evaluated under native-like and destabilizing conditions. Fibrillogenic regions were identified by bioinformatics approaches, protein fragmentation and peptide aggregation. Results. We identified four fibrillogenic regions in the HsTPI corresponding to the β3, β6, β7 y α8 of the TIM barrel. From these, the β3-strand region (residues 59-66) was highly fibrillogenic. In aggregation assays, HsTPI under native-like conditions led to amorphous assemblies while under partially denaturing conditions (urea 3.2 M) formed more structured aggregates. This slightly structured aggregates exhibited residual cross-β structure, as demonstrated by the recognition of the WO1 antibody and ATR-FTIR analysis. Discussion. Despite the fibrillogenic regions present in HsTPI, the enzyme maintained under native-favoring conditions displayed low fibrillogenic propensity. This amyloid-resistance can be attributed to the three-dimensional arrangement of the protein, where β-strands, susceptible to aggregation, are protected in the core of the molecule. Destabilization of the protein structure may expose inner regions promoting β-aggregation, as well as the formation of hydrophobic disordered aggregates. Being this last pathway kinetically favored over the thermodynamically more stable fibril aggregation pathway.
Keywords: Aggregation; Amyloid; Cross-β; Fibrillogenesis; Triosephosphate isomerase.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

Similar articles
-
Folding of beta/alpha-unit scrambled forms of S. cerevisiae triosephosphate isomerase: Evidence for autonomy of substructure formation and plasticity of hydrophobic and hydrogen bonding interactions in core of (beta/alpha)8-barrel.Proteins. 2004 May 15;55(3):548-57. doi: 10.1002/prot.20066. Proteins. 2004. PMID: 15103619
-
The CDR1 and Other Regions of Immunoglobulin Light Chains are Hot Spots for Amyloid Aggregation.Sci Rep. 2019 Feb 28;9(1):3123. doi: 10.1038/s41598-019-39781-3. Sci Rep. 2019. PMID: 30816248 Free PMC article.
-
Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro.Biochemistry. 2000 Aug 1;39(30):8735-46. doi: 10.1021/bi000276j. Biochemistry. 2000. PMID: 10913285
-
Peptide-based inhibitors of amyloid assembly.Methods Enzymol. 2006;413:273-312. doi: 10.1016/S0076-6879(06)13015-3. Methods Enzymol. 2006. PMID: 17046402 Review.
-
Structural integrity of beta-sheet assembly.Biochem Soc Trans. 2009 Aug;37(Pt 4):671-6. doi: 10.1042/BST0370671. Biochem Soc Trans. 2009. PMID: 19614573 Review.
Cited by
-
A Practical Guide to Computational Tools for Engineering Biocatalytic Properties.Int J Mol Sci. 2025 Jan 24;26(3):980. doi: 10.3390/ijms26030980. Int J Mol Sci. 2025. PMID: 39940748 Free PMC article. Review.
-
Native aggregation is a common feature among triosephosphate isomerases of different species.Sci Rep. 2020 Jan 28;10(1):1338. doi: 10.1038/s41598-020-58272-4. Sci Rep. 2020. PMID: 31992784 Free PMC article.
References
-
- Aguirre Y, Cabrera N, Aguirre B, Perez-Montfort R, Hernandez-Santoyo A, Reyes-Vivas H, Enriquez-Flores S, De Gomez-Puyou MT, Gomez-Puyou A, Sanchez-Ruiz JM, Costas M. Different contribution of conserved amino acids to the global properties of triosephosphate isomerases. Proteins. 2014;82:323–335. doi: 10.1002/prot.24398. - DOI - PubMed
-
- Bader R, Bamford R, Zurdo J, Luisi BF, Dobson CM. Probing the mechanism of amyloidogenesis through a tandem repeat of the PI3-SH3 domain suggests a generic model for protein aggregation and fibril formation. Journal of Molecular Biology. 2006;356:189–208. doi: 10.1016/j.jmb.2005.11.034. - DOI - PubMed
-
- Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, Christodoulou J, Anthony-Cahill SJ, Barker PD, Vendruscolo M, Dobson CM. Metastability of native proteins and the phenomenon of amyloid formation. Journal of the American Chemical Society. 2011;133:14160–14163. doi: 10.1021/ja2017703. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

