Metabolic Catastrophe in Mice Lacking Transferrin Receptor in Muscle
- PMID: 26870796
- PMCID: PMC4740293
- DOI: 10.1016/j.ebiom.2015.09.041
Metabolic Catastrophe in Mice Lacking Transferrin Receptor in Muscle
Abstract
Transferrin receptor (Tfr1) is ubiquitously expressed, but its roles in non-hematopoietic cells are incompletely understood. We used a tissue-specific conditional knockout strategy to ask whether skeletal muscle required Tfr1 for iron uptake. We found that iron assimilation via Tfr1 was critical for skeletal muscle metabolism, and that iron deficiency in muscle led to dramatic changes, not only in muscle, but also in adipose tissue and liver. Inactivation of Tfr1 incapacitated normal energy production in muscle, leading to growth arrest and a muted attempt to switch to fatty acid β oxidation, using up fat stores. Starvation signals stimulated gluconeogenesis in the liver, but amino acid substrates became limiting and hypoglycemia ensued. Surprisingly, the liver was also iron deficient, and production of the iron regulatory hormone hepcidin was depressed. Our observations reveal a complex interaction between iron homeostasis and metabolism that has implications for metabolic and iron disorders.
Keywords: Hepcidin; Intermediary metabolism; Iron; Skeletal muscle; Transferrin receptor.
Figures


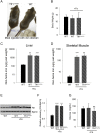
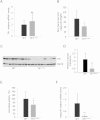
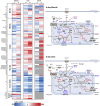

Comment in
-
Muscle specific iron deficiency has systemic consequences.EBioMedicine. 2015 Oct 5;2(11):1582-3. doi: 10.1016/j.ebiom.2015.10.001. eCollection 2015 Nov. EBioMedicine. 2015. PMID: 26870775 Free PMC article. No abstract available.
References
-
- An J., Muoio D.M., Shiota M., Fujimoto Y., Cline G.W., Shulman G.I., Koves T.R., Stevens R., Millington D., Newgard C.B. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med. 2004;10:268–274. - PubMed
-
- Andrews N.C. Medical Progress: Disorders of Iron Metabolism. N. Engl. J. Med. 1999;341:1986–1995. - PubMed
-
- Baker E., Baker S.M., Morgan E.H. Characterisation of non-transferrin-bound iron (ferric citrate) uptake by rat hepatocytes in culture. Biochim. Biophys. Acta. 1998;1380:21–30. - PubMed
-
- Barisani D., Berg C.L., Wessling-Resnick M., Gollan J.L. Evidence for a low Km transporter for non-transferrin-bound iron in isolated rat hepatocytes. Am. J. Phys. 1995;269:G570–G576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

