Human Leukemic Cells performing Oxidative Phosphorylation (OXPHOS) Generate an Antioxidant Response Independently of Reactive Oxygen species (ROS) Production
- PMID: 26870816
- PMCID: PMC4739420
- DOI: 10.1016/j.ebiom.2015.11.045
Human Leukemic Cells performing Oxidative Phosphorylation (OXPHOS) Generate an Antioxidant Response Independently of Reactive Oxygen species (ROS) Production
Abstract
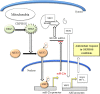
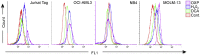
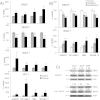
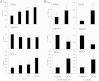
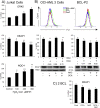
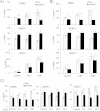
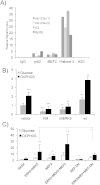
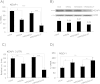
Tumor cell metabolism is altered during leukemogenesis. Cells performing oxidative phosphorylation (OXPHOS) generate reactive oxygen species (ROS) through mitochondrial activity. To limit the deleterious effects of excess ROS, certain gene promoters contain antioxidant response elements (ARE), e.g. the genes NQO-1 and HO-1. ROS induces conformational changes in KEAP1 and releases NRF2, which activates AREs. We show in vitro and in vivo that OXPHOS induces, both in primary leukemic cells and cell lines, de novo expression of NQO-1 and HO-1 and also the MAPK ERK5 and decreases KEAP1 mRNA. ERK5 activates the transcription factor MEF2, which binds to the promoter of the miR-23a-27a-24-2 cluster. Newly generated miR-23a destabilizes KEAP1 mRNA by binding to its 3'UTR. Lower KEAP1 levels increase the basal expression of the NRF2-dependent genes NQO-1 and HO-1. Hence, leukemic cells performing OXPHOS, independently of de novo ROS production, generate an antioxidant response to protect themselves from ROS.
Keywords: Antioxidant response elements (ARE); ERK5; MEF2; Mitochondria; Oxidative phosphorylation (OXPHOS); miR-23.
Figures

Comment in
-
Keep Harm at Bay: Oxidative Phosphorylation Induces Nrf2-Driven Antioxidant Response Via ERK5/MEF2/miR-23a Signaling to Keap-1.EBioMedicine. 2016 Jan 11;3:4-5. doi: 10.1016/j.ebiom.2016.01.009. eCollection 2016 Jan. EBioMedicine. 2016. PMID: 26870832 Free PMC article. No abstract available.
References
-
- Allende-Vega N., Krzywinska E., Orecchioni S., Lopez-Royuela N., Reggiani F., Talarico G., Rossi J.F., Rossignol R., Hicheri Y., Cartron G., Bertolini F., Villalba M. The presence of wild type p53 in hematological cancers improves the efficacy of combinational therapy targeting metabolism. Oncotarget. 2015;6:19228–19245. - PMC - PubMed
-
- Bellance N., Lestienne P., Rossignol R. Mitochondria: from bioenergetics to the metabolic regulation of carcinogenesis. Front. Biosci. 2009;14:4015–4034. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

