Probing the Catalytic Mechanism of Vibrio harveyi GH20 β-N-Acetylglucosaminidase by Chemical Rescue
- PMID: 26870945
- PMCID: PMC4752478
- DOI: 10.1371/journal.pone.0149228
Probing the Catalytic Mechanism of Vibrio harveyi GH20 β-N-Acetylglucosaminidase by Chemical Rescue
Abstract
Background: Vibrio harveyi GH20 β-N-acetylglucosaminidase (VhGlcNAcase) is a chitinolytic enzyme responsible for the successive degradation of chitin fragments to GlcNAc monomers, activating the onset of the chitin catabolic cascade in marine Vibrios.
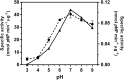
Methods: Two invariant acidic pairs (Asp303-Asp304 and Asp437-Glu438) of VhGlcNAcase were mutated using a site-directed mutagenesis strategy. The effects of these mutations were examined and the catalytic roles of these active-site residues were elucidated using a chemical rescue approach. Enhancement of the enzymic activity of the VhGlcNAcase mutants was evaluated by a colorimetric assay using pNP-GlcNAc as substrate.
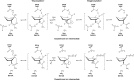
Results: Substitution of Asp303, Asp304, Asp437 or Glu438 with Ala/Asn/Gln produced a dramatic loss of the GlcNAcase activity. However, the activity of the inactive D437A mutant was recovered in the presence of sodium formate. Our kinetic data suggest that formate ion plays a nucleophilic role by mimicking the β-COO-side chain of Asp437, thereby stabilizing the reaction intermediate during both the glycosylation and the deglycosylation steps.
Conclusions: Chemical rescue of the inactive D437A mutant of VhGlcNAcase by an added nucleophile helped to identify Asp437 as the catalytic nucleophile/base, and hence its acidic partner Glu438 as the catalytic proton donor/acceptor.
General significance: Identification of the catalytic nucleophile of VhGlcNAcases supports the proposal of a substrate-assisted mechanism of GH20 GlcNAcases, requiring the catalytic pair Asp437-Glu438 for catalysis. The results suggest the mechanistic basis of the participation of β-N-acetylglucosaminidase in the chitin catabolic pathway of marine Vibrios.
Conflict of interest statement
Figures




Similar articles
-
Structural basis of chitin utilization by a GH20 β-N-acetylglucosaminidase from Vibrio campbellii strain ATCC BAA-1116.Acta Crystallogr D Struct Biol. 2021 May 1;77(Pt 5):674-689. doi: 10.1107/S2059798321002771. Epub 2021 Apr 27. Acta Crystallogr D Struct Biol. 2021. PMID: 33950022 Free PMC article.
-
Expression, purification, crystallization and preliminary crystallographic analysis of a GH20 β-N-acetylglucosaminidase from the marine bacterium Vibrio harveyi.Acta Crystallogr F Struct Biol Commun. 2015 Apr;71(Pt 4):427-33. doi: 10.1107/S2053230X1500415X. Epub 2015 Mar 20. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25849504 Free PMC article.
-
A three-step "ping-pong" mechanism of a GH20 β-N-acetylglucosaminidase from Vibrio campbellii belonging to a major Clade A-I of the phylogenetic tree of the enzyme superfamily.Biochem Biophys Res Commun. 2024 Oct 15;729:150357. doi: 10.1016/j.bbrc.2024.150357. Epub 2024 Jul 5. Biochem Biophys Res Commun. 2024. PMID: 39002194
-
Potent inhibition of a GH20 exo-β-N-acetylglucosaminidase from marine Vibrio bacteria by reaction intermediate analogues.Int J Biol Macromol. 2018 Aug;115:1165-1173. doi: 10.1016/j.ijbiomac.2018.04.193. Epub 2018 May 3. Int J Biol Macromol. 2018. PMID: 29730005
-
Enzymatic properties of β-N-acetylglucosaminidases.Appl Microbiol Biotechnol. 2018 Jan;102(1):93-103. doi: 10.1007/s00253-017-8624-7. Epub 2017 Nov 16. Appl Microbiol Biotechnol. 2018. PMID: 29143882 Review.
Cited by
-
Identification and Functional Characterization of a Novel OprD-like Chitin Uptake Channel in Non-chitinolytic Bacteria.J Biol Chem. 2016 Jun 24;291(26):13622-33. doi: 10.1074/jbc.M116.728881. Epub 2016 May 12. J Biol Chem. 2016. PMID: 27226611 Free PMC article.
-
Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.J Biol Chem. 2018 Apr 6;293(14):5150-5159. doi: 10.1074/jbc.RA117.001012. Epub 2018 Feb 14. J Biol Chem. 2018. PMID: 29444825 Free PMC article.
-
Distinctive molecular and biochemical characteristics of a glycoside hydrolase family 20 β-N-acetylglucosaminidase and salt tolerance.BMC Biotechnol. 2017 Apr 11;17(1):37. doi: 10.1186/s12896-017-0358-1. BMC Biotechnol. 2017. PMID: 28399848 Free PMC article.
-
A Shinella β-N-acetylglucosaminidase of glycoside hydrolase family 20 displays novel biochemical and molecular characteristics.Extremophiles. 2017 Jul;21(4):699-709. doi: 10.1007/s00792-017-0935-1. Epub 2017 Apr 21. Extremophiles. 2017. PMID: 28432475
-
Electrochemical N-Acetyl-β-D-glucosaminidase Urinalysis: Toward Sensor Chip-Based Diagnostics of Kidney Malfunction.Biomolecules. 2021 Sep 30;11(10):1433. doi: 10.3390/biom11101433. Biomolecules. 2021. PMID: 34680066 Free PMC article.
References
-
- Bassler BL, Yu C, Lee YC, Roseman S. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem 1991; 266:24276–24286. - PubMed
-
- Yu C, Lee AM, Bassler BL, Roseman S. Chitin utilization by marine bacteria. A physiological function for bacterial adhesion to immobilized carbohydrates. J Biol Chem 1991; 266:24260–24267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

