The Epigenomic Landscape of Prokaryotes
- PMID: 26870957
- PMCID: PMC4752239
- DOI: 10.1371/journal.pgen.1005854
The Epigenomic Landscape of Prokaryotes
Erratum in
-
Correction: The Epigenomic Landscape of Prokaryotes.PLoS Genet. 2016 May 12;12(5):e1006064. doi: 10.1371/journal.pgen.1006064. eCollection 2016 May. PLoS Genet. 2016. PMID: 27171000 Free PMC article.
Abstract
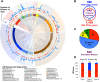
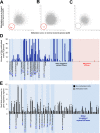
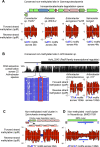
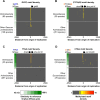
DNA methylation acts in concert with restriction enzymes to protect the integrity of prokaryotic genomes. Studies in a limited number of organisms suggest that methylation also contributes to prokaryotic genome regulation, but the prevalence and properties of such non-restriction-associated methylation systems remain poorly understood. Here, we used single molecule, real-time sequencing to map DNA modifications including m6A, m4C, and m5C across the genomes of 230 diverse bacterial and archaeal species. We observed DNA methylation in nearly all (93%) organisms examined, and identified a total of 834 distinct reproducibly methylated motifs. This data enabled annotation of the DNA binding specificities of 620 DNA Methyltransferases (MTases), doubling known specificities for previously hard to study Type I, IIG and III MTases, and revealing their extraordinary diversity. Strikingly, 48% of organisms harbor active Type II MTases with no apparent cognate restriction enzyme. These active 'orphan' MTases are present in diverse bacterial and archaeal phyla and show motif specificities and methylation patterns consistent with functions in gene regulation and DNA replication. Our results reveal the pervasive presence of DNA methylation throughout the prokaryotic kingdoms, as well as the diversity of sequence specificities and potential functions of DNA methylation systems.
Conflict of interest statement
RJR, RDM, JP and AF are employees of New England Biolabs (NEB), a company that sells DNA methylases and restriction enzymes. NEB paid for a part of the research. JK and TAC are full-time employees at Pacific Biosciences, a company commercializing single molecule, real-time sequencing technologies.
Figures


Comment in
-
DNA methylation: A SMRT analysis of prokaryotic epigenomes.Nat Rev Genet. 2016 Apr;17(4):195. doi: 10.1038/nrg.2016.22. Epub 2016 Feb 29. Nat Rev Genet. 2016. PMID: 26924764 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

