Networking in the Plant Microbiome
- PMID: 26871440
- PMCID: PMC4752285
- DOI: 10.1371/journal.pbio.1002378
Networking in the Plant Microbiome
Abstract
Almost all higher organisms, including plants, insects, and mammals, are colonized by complex microbial communities and harbor a microbiome. Emerging studies with plants reveal that these microbiomes are structured and form complex, interconnected microbial networks. Within these networks, different taxa have different roles, and keystone species have been identified that could be crucial for plant health and ecosystem functioning. A new paper in this issue of PLOS Biology by Agler et al. highlights the presence of microbial hubs in these networks that may act as mediators between the plant and its microbiome. A next major frontier is now to link microbiome composition to function. In order to do this, we present a number of hypothetical examples of how microbiome diversity and function potentially influence host performance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

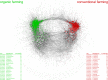
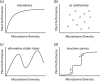
Comment on
-
Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation.PLoS Biol. 2016 Jan 20;14(1):e1002352. doi: 10.1371/journal.pbio.1002352. eCollection 2016 Jan. PLoS Biol. 2016. PMID: 26788878 Free PMC article.
References
-
- Turnbaugh P, Ley R, Mahowald M, Magrini V, Mardis E, Gordon J. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. - PubMed
-
- Dillon R, Dillon V. The gut bacteria of insects: nonpathogenic interactions. Annual Reviews in Entomology. 2004;49(1):71–92. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

