Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells
- PMID: 26871630
- PMCID: PMC4752721
- DOI: 10.1016/j.cell.2015.12.039
Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells
Abstract
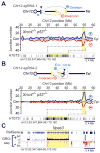
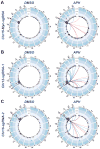
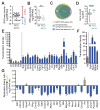
Repair of DNA double-strand breaks (DSBs) by non-homologous end joining is critical for neural development, and brain cells frequently contain somatic genomic variations that might involve DSB intermediates. We now use an unbiased, high-throughput approach to identify genomic regions harboring recurrent DSBs in primary neural stem/progenitor cells (NSPCs). We identify 27 recurrent DSB clusters (RDCs), and remarkably, all occur within gene bodies. Most of these NSPC RDCs were detected only upon mild, aphidicolin-induced replication stress, providing a nucleotide-resolution view of replication-associated genomic fragile sites. The vast majority of RDCs occur in long, transcribed, and late-replicating genes. Moreover, almost 90% of identified RDC-containing genes are involved in synapse function and/or neural cell adhesion, with a substantial fraction also implicated in tumor suppression and/or mental disorders. Our characterization of NSPC RDCs reveals a basis of gene fragility and suggests potential impacts of DNA breaks on neurodevelopment and neural functions.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
A Mechanism for Somatic Brain Mosaicism.Cell. 2016 Feb 11;164(4):593-5. doi: 10.1016/j.cell.2016.01.048. Cell. 2016. PMID: 26871622
-
Molecular biology: Breaks in the brain.Nature. 2016 Apr 7;532(7597):46-7. doi: 10.1038/nature17316. Epub 2016 Mar 23. Nature. 2016. PMID: 27007850 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

