Development of Antibiotic Resistance during Simulated Treatment of Pseudomonas aeruginosa in Chemostats
- PMID: 26872140
- PMCID: PMC4752458
- DOI: 10.1371/journal.pone.0149310
Development of Antibiotic Resistance during Simulated Treatment of Pseudomonas aeruginosa in Chemostats
Abstract
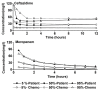
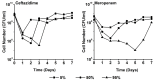
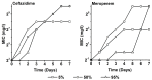
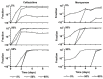
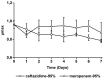
During treatment of infections with antibiotics in critically ill patients in the intensive care resistance often develops. This study aims to establish whether under those conditions this resistance can develop de novo or that genetic exchange between bacteria is by necessity involved. Chemostat cultures of Pseudomonas aeruginosa were exposed to treatment regimes with ceftazidime and meropenem that simulated conditions expected in patient plasma. Development of antibiotic resistance was monitored and mutations in resistance genes were searched for by sequencing PCR products. Even at the highest concentrations that can be expected in patients, sufficient bacteria survived in clumps of filamentous cells to recover and grow out after 3 to 5 days. At the end of a 7 days simulated treatment, the minimal inhibitory concentration (MIC) had increased by a factor between 10 and 10,000 depending on the antibiotic and the treatment protocol. The fitness costs of resistance were minimal. In the resistant strains, only three mutations were observed in genes associated with beta-lactam resistance. The development of resistance often observed during patient treatment can be explained by de novo acquisition of resistance and genetic exchange of resistance genes is not by necessity involved. As far as conclusions based on an in vitro study using P. aeruginosa and only two antibiotics can be generalized, it seems that development of resistance can be minimized by treating with antibiotics in the highest concentration the patient can endure for the shortest time needed to eliminate the infection.
Conflict of interest statement
Figures

Similar articles
-
Optimization of therapy against Pseudomonas aeruginosa with ceftazidime and meropenem using chemostats as model for infections.FEMS Microbiol Lett. 2017 Aug 1;364(14). doi: 10.1093/femsle/fnx142. FEMS Microbiol Lett. 2017. PMID: 28854670
-
In-vitro effects of a combination of antipseudomonal antibiotics against multi-drug resistant Pseudomonas aeruginosa.J Antimicrob Chemother. 1999 Nov;44(5):689-91. doi: 10.1093/jac/44.5.689. J Antimicrob Chemother. 1999. PMID: 10552987
-
Development of resistance in wild-type and hypermutable Pseudomonas aeruginosa strains exposed to clinical pharmacokinetic profiles of meropenem and ceftazidime simulated in vitro.Antimicrob Agents Chemother. 2007 Oct;51(10):3642-9. doi: 10.1128/AAC.00160-07. Epub 2007 Aug 6. Antimicrob Agents Chemother. 2007. PMID: 17682103 Free PMC article.
-
Activity of Ceftolozane-Tazobactam and Ceftazidime-Avibactam against Beta-Lactam-Resistant Pseudomonas aeruginosa Isolates.Antimicrob Agents Chemother. 2017 Nov 22;61(12):e01858-17. doi: 10.1128/AAC.01858-17. Print 2017 Dec. Antimicrob Agents Chemother. 2017. PMID: 28993338 Free PMC article.
-
Pseudomonas aeruginosa chromosomal beta-lactamase in patients with cystic fibrosis and chronic lung infection. Mechanism of antibiotic resistance and target of the humoral immune response.APMIS Suppl. 2003;(116):1-47. APMIS Suppl. 2003. PMID: 14692154 Review.
Cited by
-
Antibiotic selection in the treatment of acute invasive infections by Pseudomonas aeruginosa: Guidelines by the Spanish Society of Chemotherapy.Rev Esp Quimioter. 2018 Feb;31(1):78-100. Epub 2018 Feb 23. Rev Esp Quimioter. 2018. PMID: 29480677 Free PMC article. Review.
-
Adaptive response of Pseudomonas aeruginosa under serial ciprofloxacin exposure.Microbiology (Reading). 2024 Apr;170(3):001443. doi: 10.1099/mic.0.001443. Microbiology (Reading). 2024. PMID: 38568202 Free PMC article.
-
Recent Trends in S. aureus and E. coli-Based Endometritis, and the Therapeutic Evaluation of Sodium Alginate-Based Antibiotics and Nanoparticles.Gels. 2023 Dec 5;9(12):955. doi: 10.3390/gels9120955. Gels. 2023. PMID: 38131941 Free PMC article.
-
Unlocking the potential of experimental evolution to study drug resistance in pathogenic fungi.NPJ Antimicrob Resist. 2024 Dec 12;2(1):48. doi: 10.1038/s44259-024-00064-1. NPJ Antimicrob Resist. 2024. PMID: 39843963 Free PMC article. Review.
-
Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria?GMS Hyg Infect Control. 2017 Apr 10;12:Doc05. doi: 10.3205/dgkh000290. eCollection 2017. GMS Hyg Infect Control. 2017. PMID: 28451516 Free PMC article.
References
-
- Gaieski DF, Mikkelsen ME, Band RA, Pines JM, Massone R, Furia FF, et al. (2010) Impact of time to antibiotics on survival in patients with severe sepsis or septic shock in whom early goal-directed therapy was initiated in the emergency department. Crit Care Med 38: 1045–1053. 10.1097/CCM.0b013e3181cc4824 - DOI - PubMed
-
- Garnacho-Montero J, Garcia-Garmendia JL, Barrero-Almodovar A, Jimenez-Jimenez FJ, Perez-Paredes C, Ortiz-Leyba C (2003) Impact of adequate empirical antibiotic therapy on the outcome of patients admitted to the intensive care unit with sepsis. Crit Care Med 31: 2742–2751. - PubMed
-
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, et al. (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34: 1589–1596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

