Neighborhood Regularized Logistic Matrix Factorization for Drug-Target Interaction Prediction
- PMID: 26872142
- PMCID: PMC4752318
- DOI: 10.1371/journal.pcbi.1004760
Neighborhood Regularized Logistic Matrix Factorization for Drug-Target Interaction Prediction
Abstract
In pharmaceutical sciences, a crucial step of the drug discovery process is the identification of drug-target interactions. However, only a small portion of the drug-target interactions have been experimentally validated, as the experimental validation is laborious and costly. To improve the drug discovery efficiency, there is a great need for the development of accurate computational approaches that can predict potential drug-target interactions to direct the experimental verification. In this paper, we propose a novel drug-target interaction prediction algorithm, namely neighborhood regularized logistic matrix factorization (NRLMF). Specifically, the proposed NRLMF method focuses on modeling the probability that a drug would interact with a target by logistic matrix factorization, where the properties of drugs and targets are represented by drug-specific and target-specific latent vectors, respectively. Moreover, NRLMF assigns higher importance levels to positive observations (i.e., the observed interacting drug-target pairs) than negative observations (i.e., the unknown pairs). Because the positive observations are already experimentally verified, they are usually more trustworthy. Furthermore, the local structure of the drug-target interaction data has also been exploited via neighborhood regularization to achieve better prediction accuracy. We conducted extensive experiments over four benchmark datasets, and NRLMF demonstrated its effectiveness compared with five state-of-the-art approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



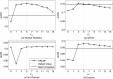
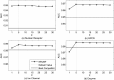



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

