Localization of the binding interface between leiomodin-2 and α-tropomyosin
- PMID: 26873245
- PMCID: PMC5079754
- DOI: 10.1016/j.bbapap.2016.02.009
Localization of the binding interface between leiomodin-2 and α-tropomyosin
Abstract
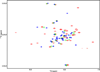
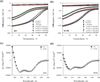
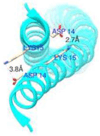
The development of some familial dilated cardiomyopathies (DCM) correlates with the presence of mutations in proteins that regulate the organization and function of thin filaments in cardiac muscle cells. Harmful effects of some mutations might be caused by disruption of yet uncharacterized protein-protein interactions. We used nuclear magnetic resonance spectroscopy to localize the region of striated muscle α-tropomyosin (Tpm1.1) that interacts with leiomodin-2 (Lmod2), a member of tropomodulin (Tmod) family of actin-binding proteins. We found that 21 N-terminal residues of Tpm1.1 are involved in interactions with residues 7-41 of Lmod2. The K15N mutation in Tpm1.1, known to be associated with familial DCM, is located within the newly identified Lmod2 binding site of Tpm1.1. We studied the effect of this mutation on binding Lmod2 and Tmod1. The mutation reduced binding affinity for both Lmod2 and Tmod1, which are responsible for correct lengths of thin filaments. The effect of the K15N mutation on Tpm1.1 binding to Lmod2 and Tmod1 provides a molecular rationale for the development of familial DCM.
Keywords: Circular dichroism; Dilated cardiomyopathy; Intrinsically disordered regions; Leiomodin; Nuclear magnetic resonance; Tropomodulin.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures

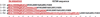


Similar articles
-
The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2's function.Mol Biol Cell. 2016 Aug 15;27(16):2565-75. doi: 10.1091/mbc.E16-03-0200. Epub 2016 Jun 15. Mol Biol Cell. 2016. PMID: 27307584 Free PMC article.
-
The cardiomyopathy-associated K15N mutation in tropomyosin alters actin filament pointed end dynamics.Arch Biochem Biophys. 2017 Sep 15;630:18-26. doi: 10.1016/j.abb.2017.07.006. Epub 2017 Jul 18. Arch Biochem Biophys. 2017. PMID: 28732641 Free PMC article.
-
Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle.J Cell Sci. 2010 Sep 15;123(Pt 18):3136-45. doi: 10.1242/jcs.071837. Epub 2010 Aug 24. J Cell Sci. 2010. PMID: 20736303 Free PMC article.
-
Tropomodulin capping of actin filaments in striated muscle development and physiology.J Biomed Biotechnol. 2011;2011:103069. doi: 10.1155/2011/103069. Epub 2011 Oct 17. J Biomed Biotechnol. 2011. PMID: 22013379 Free PMC article. Review.
-
Tropomodulins and tropomyosins: working as a team.J Muscle Res Cell Motil. 2013 Aug;34(3-4):247-60. doi: 10.1007/s10974-013-9349-6. Epub 2013 Jul 5. J Muscle Res Cell Motil. 2013. PMID: 23828180 Free PMC article. Review.
Cited by
-
Effects of cardiomyopathy-linked mutations K15N and R21H in tropomyosin on thin-filament regulation and pointed-end dynamics.Mol Biol Cell. 2019 Jan 15;30(2):268-281. doi: 10.1091/mbc.E18-06-0406. Epub 2018 Nov 21. Mol Biol Cell. 2019. PMID: 30462572 Free PMC article.
-
The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2's function.Mol Biol Cell. 2016 Aug 15;27(16):2565-75. doi: 10.1091/mbc.E16-03-0200. Epub 2016 Jun 15. Mol Biol Cell. 2016. PMID: 27307584 Free PMC article.
-
Cardiac leiomodin2 binds to the sides of actin filaments and regulates the ATPase activity of myosin.PLoS One. 2017 Oct 12;12(10):e0186288. doi: 10.1371/journal.pone.0186288. eCollection 2017. PLoS One. 2017. PMID: 29023566 Free PMC article.
-
Prediction and biological significance of small changes in binding of leiomodin to tropomyosin.J Gen Physiol. 2025 Jul 7;157(4):e202413641. doi: 10.1085/jgp.202413641. Epub 2025 Apr 24. J Gen Physiol. 2025. PMID: 40272387
-
Characterizing interaction forces between actin and proteins of the tropomodulin family reveals the presence of the N-terminal actin-binding site in leiomodin.Arch Biochem Biophys. 2018 Jan 15;638:18-26. doi: 10.1016/j.abb.2017.12.005. Epub 2017 Dec 6. Arch Biochem Biophys. 2018. PMID: 29223925 Free PMC article.
References
-
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. Journal of cell science. 2015;128:2965–2974. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

