Use of Metagenomic Shotgun Sequencing Technology To Detect Foodborne Pathogens within the Microbiome of the Beef Production Chain
- PMID: 26873315
- PMCID: PMC4959480
- DOI: 10.1128/AEM.00078-16
Use of Metagenomic Shotgun Sequencing Technology To Detect Foodborne Pathogens within the Microbiome of the Beef Production Chain
Abstract
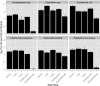
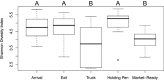
Foodborne illnesses associated with pathogenic bacteria are a global public health and economic challenge. The diversity of microorganisms (pathogenic and nonpathogenic) that exists within the food and meat industries complicates efforts to understand pathogen ecology. Further, little is known about the interaction of pathogens within the microbiome throughout the meat production chain. Here, a metagenomic approach and shotgun sequencing technology were used as tools to detect pathogenic bacteria in environmental samples collected from the same groups of cattle at different longitudinal processing steps of the beef production chain: cattle entry to feedlot, exit from feedlot, cattle transport trucks, abattoir holding pens, and the end of the fabrication system. The log read counts classified as pathogens per million reads for Salmonella enterica,Listeria monocytogenes,Escherichia coli,Staphylococcus aureus, Clostridium spp. (C. botulinum and C. perfringens), and Campylobacter spp. (C. jejuni,C. coli, and C. fetus) decreased over subsequential processing steps. Furthermore, the normalized read counts for S. enterica,E. coli, and C. botulinumwere greater in the final product than at the feedlots, indicating that the proportion of these bacteria increased (the effect on absolute numbers was unknown) within the remaining microbiome. From an ecological perspective, data indicated that shotgun metagenomics can be used to evaluate not only the microbiome but also shifts in pathogen populations during beef production. Nonetheless, there were several challenges in this analysis approach, one of the main ones being the identification of the specific pathogen from which the sequence reads originated, which makes this approach impractical for use in pathogen identification for regulatory and confirmation purposes.
Copyright © 2016 Yang et al.
Figures




Similar articles
-
Shotgun-metagenomics reveals a highly diverse and communal microbial network present in the drains of three beef-processing plants.Front Cell Infect Microbiol. 2023 Sep 8;13:1240138. doi: 10.3389/fcimb.2023.1240138. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37743870 Free PMC article.
-
Microbial dynamics of South Korean beef and surroundings along the supply chain based on high-throughput sequencing.Meat Sci. 2024 Aug;214:109520. doi: 10.1016/j.meatsci.2024.109520. Epub 2024 Apr 24. Meat Sci. 2024. PMID: 38703561
-
Evaluating the Prevalence of Foodborne Pathogens in Livestock Using Metagenomics Approach.J Microbiol Biotechnol. 2021 Dec 28;31(12):1701-1708. doi: 10.4014/jmb.2109.09038. J Microbiol Biotechnol. 2021. PMID: 34675137 Free PMC article.
-
The diversity of beef safety: A global reason to strengthen our current systems.Meat Sci. 2017 Oct;132:59-71. doi: 10.1016/j.meatsci.2017.03.015. Epub 2017 Apr 5. Meat Sci. 2017. PMID: 28434613 Review.
-
Sodium chloride reduction in meat processing: Microbial shifts, spoilage risks, and metagenomic insights.Meat Sci. 2025 Aug;226:109848. doi: 10.1016/j.meatsci.2025.109848. Epub 2025 May 10. Meat Sci. 2025. PMID: 40373419 Review.
Cited by
-
Precise Species Detection in Traditional Herbal Patent Medicine, Qingguo Wan, Using Shotgun Metabarcoding.Front Pharmacol. 2021 Apr 28;12:607210. doi: 10.3389/fphar.2021.607210. eCollection 2021. Front Pharmacol. 2021. PMID: 33995010 Free PMC article.
-
Significant loss of sensitivity and specificity in the taxonomic classification occurs when short 16S rRNA gene sequences are used.Heliyon. 2016 Sep 23;2(9):e00170. doi: 10.1016/j.heliyon.2016.e00170. eCollection 2016 Sep. Heliyon. 2016. PMID: 27699286 Free PMC article.
-
Liver abscess microbiota of feedlot steers finished in natural and traditional management programs.J Anim Sci. 2022 Nov 1;100(11):skac252. doi: 10.1093/jas/skac252. J Anim Sci. 2022. PMID: 35938914 Free PMC article.
-
Shotgun-metagenomics reveals a highly diverse and communal microbial network present in the drains of three beef-processing plants.Front Cell Infect Microbiol. 2023 Sep 8;13:1240138. doi: 10.3389/fcimb.2023.1240138. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37743870 Free PMC article.
-
Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems.Sci Rep. 2016 Apr 20;6:24645. doi: 10.1038/srep24645. Sci Rep. 2016. PMID: 27095377 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

