Expression Profiling of Macrophages Reveals Multiple Populations with Distinct Biological Roles in an Immunocompetent Orthotopic Model of Lung Cancer
- PMID: 26873985
- PMCID: PMC4779748
- DOI: 10.4049/jimmunol.1502364
Expression Profiling of Macrophages Reveals Multiple Populations with Distinct Biological Roles in an Immunocompetent Orthotopic Model of Lung Cancer
Abstract
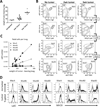
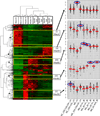
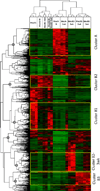
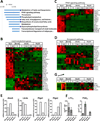
Macrophages represent an important component of the tumor microenvironment and play a complex role in cancer progression. These cells are characterized by a high degree of plasticity, and they alter their phenotype in response to local environmental cues. Whereas the M1/M2 classification of macrophages has been widely used, the complexity of macrophage phenotypes has not been well studied, particularly in lung cancer. In this study we employed an orthotopic immunocompetent model of lung adenocarcinoma in which murine lung cancer cells are directly implanted into the left lobe of syngeneic mice. Using multimarker flow cytometry, we defined and recovered several distinct populations of monocytes/macrophages from tumors at different stages of progression. We used RNA-seq transcriptional profiling to define distinct features of each population and determine how they change during tumor progression. We defined an alveolar resident macrophage population that does not change in number and expresses multiple genes related to lipid metabolism and lipid signaling. We also defined a population of tumor-associated macrophages that increase dramatically with tumor and selectively expresses a panel of chemokine genes. A third population, which resembles tumor-associated monocytes, expresses a large number of genes involved in matrix remodeling. By correlating transcriptional profiles with clinically prognostic genes, we show that specific monocyte/macrophage populations are enriched in genes that predict outcomes in lung adenocarcinoma, implicating these subpopulations as critical determinants of patient survival. Our data underscore the complexity of monocytes/macrophages in the tumor microenvironment, and they suggest that distinct populations play specific roles in tumor progression.
Copyright © 2016 by The American Association of Immunologists, Inc.
Figures




References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Sharma SK, Chintala NK, Vadrevu SK, Patel J, Karbowniczek M, Markiewski MM. Pulmonary alveolar macrophages contribute to the premetastatic niche by suppressing antitumor T cell responses in the lungs. J Immunol. 2015;194:5529–5538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

