Hydroxylation and translational adaptation to stress: some answers lie beyond the STOP codon
- PMID: 26874685
- PMCID: PMC11108485
- DOI: 10.1007/s00018-016-2160-y
Hydroxylation and translational adaptation to stress: some answers lie beyond the STOP codon
Abstract
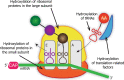
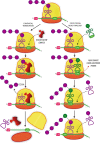
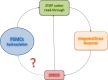
Regulation of protein synthesis contributes to maintenance of homeostasis and adaptation to environmental changes. mRNA translation is controlled at various levels including initiation, elongation and termination, through post-transcriptional/translational modifications of components of the protein synthesis machinery. Recently, protein and RNA hydroxylation have emerged as important enzymatic modifications of tRNAs, elongation and termination factors, as well as ribosomal proteins. These modifications enable a correct STOP codon recognition, ensuring translational fidelity. Recent studies are starting to show that STOP codon read-through is related to the ability of the cell to cope with different types of stress, such as oxidative and chemical insults, while correlations between defects in hydroxylation of protein synthesis components and STOP codon read-through are beginning to emerge. In this review we will discuss our current knowledge of protein synthesis regulation through hydroxylation of components of the translation machinery, with special focus on STOP codon recognition. We speculate on the possibility that programmed STOP codon read-through, modulated by hydroxylation of components of the protein synthesis machinery, is part of a concerted cellular response to stress.
Keywords: Dioxygenases; Post-translational modifications; Protein synthesis; Translational fidelity.
Figures
Similar articles
-
Structural Basis for Translation Termination on a Pseudouridylated Stop Codon.J Mol Biol. 2016 May 22;428(10 Pt B):2228-36. doi: 10.1016/j.jmb.2016.04.018. Epub 2016 Apr 20. J Mol Biol. 2016. PMID: 27107638 Free PMC article.
-
uS3/Rps3 controls fidelity of translation termination and programmed stop codon readthrough in co-operation with eIF3.Nucleic Acids Res. 2019 Dec 2;47(21):11326-11343. doi: 10.1093/nar/gkz929. Nucleic Acids Res. 2019. PMID: 31642471 Free PMC article.
-
Recognition of 3' nucleotide context and stop codon readthrough are determined during mRNA translation elongation.J Biol Chem. 2022 Jul;298(7):102133. doi: 10.1016/j.jbc.2022.102133. Epub 2022 Jun 11. J Biol Chem. 2022. PMID: 35700825 Free PMC article.
-
Dynamic basis of fidelity and speed in translation: Coordinated multistep mechanisms of elongation and termination.Protein Sci. 2017 Jul;26(7):1352-1362. doi: 10.1002/pro.3190. Epub 2017 May 23. Protein Sci. 2017. PMID: 28480640 Free PMC article. Review.
-
Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence.RNA Biol. 2015;12(9):950-8. doi: 10.1080/15476286.2015.1068497. RNA Biol. 2015. PMID: 26176195 Free PMC article. Review.
Cited by
-
The non-stop decay mRNA surveillance pathway is required for oxidative stress tolerance.Nucleic Acids Res. 2017 Jun 20;45(11):6881-6893. doi: 10.1093/nar/gkx306. Nucleic Acids Res. 2017. PMID: 28472342 Free PMC article.
-
Relocalization of Translation Termination and Ribosome Recycling Factors to Stress Granules Coincides with Elevated Stop-Codon Readthrough and Reinitiation Rates upon Oxidative Stress.Cells. 2023 Jan 8;12(2):259. doi: 10.3390/cells12020259. Cells. 2023. PMID: 36672194 Free PMC article.
-
Uniqueness of RNA Coliphage Qβ Display System in Directed Evolutionary Biotechnology.Viruses. 2021 Mar 27;13(4):568. doi: 10.3390/v13040568. Viruses. 2021. PMID: 33801772 Free PMC article. Review.
-
Nonsense Suppression as an Approach to Treat Lysosomal Storage Diseases.Diseases. 2016 Dec;4(4):32. doi: 10.3390/diseases4040032. Epub 2016 Oct 19. Diseases. 2016. PMID: 28367323 Free PMC article.
-
Oxidative Damage of Mussels Living in Seawater Enriched with Trace Metals, from the Viewpoint of Proteins Expression and Modification.Toxics. 2020 Oct 18;8(4):89. doi: 10.3390/toxics8040089. Toxics. 2020. PMID: 33081042 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

