Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158
- PMID: 26875695
- PMCID: PMC4753449
- DOI: 10.1038/srep20915
Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158
Abstract
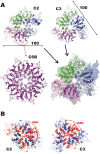
DNA replication initiation is a vital and tightly regulated step in all replicons and requires an initiator factor that specifically recognizes the DNA replication origin and starts replication. RepB from the promiscuous streptococcal plasmid pMV158 is a hexameric ring protein evolutionary related to viral initiators. Here we explore the conformational plasticity of the RepB hexamer by i) SAXS, ii) sedimentation experiments, iii) molecular simulations and iv) X-ray crystallography. Combining these techniques, we derive an estimate of the conformational ensemble in solution showing that the C-terminal oligomerisation domains of the protein form a rigid cylindrical scaffold to which the N-terminal DNA-binding/catalytic domains are attached as highly flexible appendages, featuring multiple orientations. In addition, we show that the hinge region connecting both domains plays a pivotal role in the observed plasticity. Sequence comparisons and a literature survey show that this hinge region could exists in other initiators, suggesting that it is a common, crucial structural element for DNA binding and manipulation.
Figures





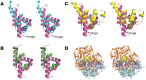
Similar articles
-
Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.Nucleic Acids Res. 2023 Feb 22;51(3):1458-1472. doi: 10.1093/nar/gkac1271. Nucleic Acids Res. 2023. PMID: 36688326 Free PMC article.
-
Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions.J Mol Biol. 1990 May 20;213(2):247-62. doi: 10.1016/S0022-2836(05)80188-3. J Mol Biol. 1990. PMID: 2160544
-
Translation initiation of the replication initiator repB gene of promiscuous plasmid pMV158 is led by an extended non-SD sequence.Plasmid. 2013 Jul;70(1):69-77. doi: 10.1016/j.plasmid.2013.01.011. Epub 2013 Feb 16. Plasmid. 2013. PMID: 23419647
-
Bringing them together: plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective.Plasmid. 2014 Jul;74:15-31. doi: 10.1016/j.plasmid.2014.05.004. Epub 2014 Jun 2. Plasmid. 2014. PMID: 24942190 Free PMC article. Review.
-
Origin inactivation in bacterial DNA replication control.Mol Microbiol. 2006 Jul;61(1):9-15. doi: 10.1111/j.1365-2958.2006.05229.x. Mol Microbiol. 2006. PMID: 16824091 Review.
Cited by
-
Successful Establishment of Plasmids R1 and pMV158 in a New Host Requires the Relief of the Transcriptional Repression of Their Essential rep Genes.Front Microbiol. 2017 Dec 1;8:2367. doi: 10.3389/fmicb.2017.02367. eCollection 2017. Front Microbiol. 2017. PMID: 29250051 Free PMC article.
-
Acidic pH Decreases the Endonuclease Activity of Initiator RepB and Increases the Stability of the Covalent RepB-DNA Intermediate while Has Only a Limited Effect on the Replication of Plasmid pMV158 in Lactococcus lactis.Front Mol Biosci. 2021 Mar 5;8:634461. doi: 10.3389/fmolb.2021.634461. eCollection 2021. Front Mol Biosci. 2021. PMID: 33889596 Free PMC article.
-
The Different Faces of Rolling-Circle Replication and Its Multifunctional Initiator Proteins.Front Microbiol. 2017 Nov 30;8:2353. doi: 10.3389/fmicb.2017.02353. eCollection 2017. Front Microbiol. 2017. PMID: 29250047 Free PMC article. Review.
-
Evidence of in vitro mecB-mediated β-lactam antibiotic resistance transfer to Staphylococcus aureus from Macrococcus psychrotolerans sp. nov., a psychrophilic bacterium from food-producing animals and human clinical specimens.Appl Environ Microbiol. 2025 Apr 23;91(4):e0165224. doi: 10.1128/aem.01652-24. Epub 2025 Mar 11. Appl Environ Microbiol. 2025. PMID: 40066988 Free PMC article.
-
Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.Nucleic Acids Res. 2023 Feb 22;51(3):1458-1472. doi: 10.1093/nar/gkac1271. Nucleic Acids Res. 2023. PMID: 36688326 Free PMC article.
References
-
- de la Campa A. G., del Solar G. H. & Espinosa M. Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J Mol Biol 213, 247–262 (1990). - PubMed
-
- Ruiz-Masó J. A., López-Zumel C., Menéndez M., Espinosa M. & del Solar G. Structural features of the initiator of replication protein RepB encoded by the promiscuous plasmid pMV158. Biochim Biophys Acta 1696, 113–119 (2004). - PubMed
-
- Dyda F. & Hickman A. B. A mob of reps. Structure 11, 1310–1311 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

