Structure and Engineering of Francisella novicida Cas9
- PMID: 26875867
- PMCID: PMC4899972
- DOI: 10.1016/j.cell.2016.01.039
Structure and Engineering of Francisella novicida Cas9
Abstract
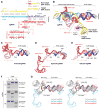
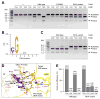
The RNA-guided endonuclease Cas9 cleaves double-stranded DNA targets complementary to the guide RNA and has been applied to programmable genome editing. Cas9-mediated cleavage requires a protospacer adjacent motif (PAM) juxtaposed with the DNA target sequence, thus constricting the range of targetable sites. Here, we report the 1.7 Å resolution crystal structures of Cas9 from Francisella novicida (FnCas9), one of the largest Cas9 orthologs, in complex with a guide RNA and its PAM-containing DNA targets. A structural comparison of FnCas9 with other Cas9 orthologs revealed striking conserved and divergent features among distantly related CRISPR-Cas9 systems. We found that FnCas9 recognizes the 5'-NGG-3' PAM, and used the structural information to create a variant that can recognize the more relaxed 5'-YG-3' PAM. Furthermore, we demonstrated that the FnCas9-ribonucleoprotein complex can be microinjected into mouse zygotes to edit endogenous sites with the 5'-YG-3' PAM, thus expanding the target space of the CRISPR-Cas9 toolbox.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





References
-
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

