Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains
- PMID: 26876100
- PMCID: PMC4775417
- DOI: 10.1016/j.str.2015.12.017
Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains
Abstract
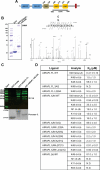
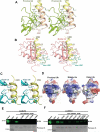
Lys48-linked ubiquitin chains act as the main targeting signals for protein degradation by the proteasome. Here we report selective binding of AIRAPL, a protein that associates with the proteasome upon exposure to arsenite, to Lys48-linked tri-ubiquitin chains. AIRAPL comprises two ubiquitin-interacting motifs in tandem (tUIMs) that are linked through a flexible inter-UIM region. In the complex crystal structure UIM1 binds the proximal ubiquitin, whereas UIM2 (the double-sided UIM) binds non-symmetrically to the middle and distal ubiquitin moieties on either side of the helix. Specificity of AIRAPL for Lys48-linked ubiquitin chains is determined by UIM2, and the flexible inter-UIM linker increases avidity by placing the two UIMs in an orientation that facilitates binding of the third ubiquitin to UIM1. Unlike middle and proximal ubiquitins, distal ubiquitin binds UIM2 through a novel surface, which leaves the Ile44 hydrophobic patch accessible for binding to the proteasomal ubiquitin receptors.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures


References
-
- CHAU V, TOBIAS JW, BACHMAIR A, MARRIOTT D, ECKER DJ, GONDA DK, VARSHAVSKY A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83. - PubMed
-
- CIECHANOVER A, STANHILL A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim Biophys Acta. 2014;1843:86–96. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

