The Apoptotic Engulfment Machinery Regulates Axonal Degeneration in C. elegans Neurons
- PMID: 26876181
- PMCID: PMC4821572
- DOI: 10.1016/j.celrep.2016.01.050
The Apoptotic Engulfment Machinery Regulates Axonal Degeneration in C. elegans Neurons
Abstract
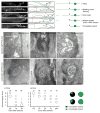
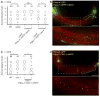
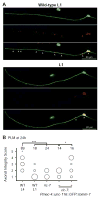
Axonal degeneration is a characteristic feature of neurodegenerative disease and nerve injury. Here, we characterize axonal degeneration in Caenorhabditis elegans neurons following laser-induced axotomy. We show that this process proceeds independently of the WLD(S) and Nmnat pathway and requires the axonal clearance machinery that includes the conserved transmembrane receptor CED-1/Draper, the adaptor protein CED-6, the guanine nucleotide exchange factor complex Crk/Mbc/dCed-12 (CED-2/CED-5/CED-12), and the small GTPase Rac1 (CED-10). We demonstrate that CED-1 and CED-6 function non-cell autonomously in the surrounding hypodermis, which we show acts as the engulfing tissue for the severed axon. Moreover, we establish a function in this process for CED-7, an ATP-binding cassette (ABC) transporter, and NRF-5, a lipid-binding protein, both associated with release of lipid-vesicles during apoptotic cell clearance. Thus, our results reveal the existence of a WLD(S)/Nmnat-independent axonal degeneration pathway, conservation of the axonal clearance machinery, and a function for CED-7 and NRF-5 in this process.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Adalbert R, Gillingwater TH, Haley JE, Bridge K, Beirowski B, Berek L, Wagner D, Grumme D, Thomson D, Celik A, et al. A rat model of slow Wallerian degeneration (WldS) with improved preservation of neuromuscular synapses. Eur J Neurosci. 2005;21:271–277. - PubMed
-
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. - PubMed
-
- Beirowski B, Babetto E, Coleman MP, Martin KR. The WldS gene delays axonal but not somatic degeneration in a rat glaucoma model. Eur J Neurosci. 2008;28:1166–1179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

