Genome-wide Analysis Identifies Bcl6-Controlled Regulatory Networks during T Follicular Helper Cell Differentiation
- PMID: 26876184
- PMCID: PMC4975778
- DOI: 10.1016/j.celrep.2016.01.038
Genome-wide Analysis Identifies Bcl6-Controlled Regulatory Networks during T Follicular Helper Cell Differentiation
Abstract
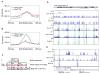
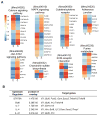
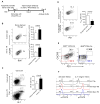
T follicular helper (Tfh) cell is a unique T cell subset specialized in promoting humoral immunity. B-cell lymphoma 6 protein (Bcl6) has been identified as an obligatory transcription factor in Tfh cells; however, the molecular mechanism underlying Bcl6 function remains largely unknown. Here, we defined Bcl6 target genes in Tfh cells by analyzing genome-wide Bcl6 occupancy together with transcriptome profiling. With consensus sequences being different from those in Th9, B cells, and macrophages, Bcl6 binding in Tfh cell was closely associated with a decrease in 5-hydroxymethylcytosine (5hmC). Importantly, Bcl6 promoted Tfh cell differentiation through antagonizing IL-7R (CD127)/signal transducer and activator of transcription (STAT) 5 axis; deletion of the Bcl6 gene in T cells resulted in enhanced IL-7R-STAT5 signaling and substantial expansion of CD127(hi) non-Tfh cells. Thus, our study systemically examines Bcl6-controlled regulatory networks and provides important insights into Bcl6's biological functions in Tfh cells.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Similar articles
-
CD25(+) Bcl6(low) T follicular helper cells provide help to maturing B cells in germinal centers of human tonsil.Eur J Immunol. 2015 Jan;45(1):298-308. doi: 10.1002/eji.201444911. Epub 2014 Oct 27. Eur J Immunol. 2015. PMID: 25263533 Free PMC article.
-
Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation.J Immunol. 2012 Apr 15;188(8):3734-44. doi: 10.4049/jimmunol.1103246. Epub 2012 Mar 16. J Immunol. 2012. PMID: 22427637 Free PMC article.
-
Repression of miR-31 by BCL6 stabilizes the helper function of human follicular helper T cells.Proc Natl Acad Sci U S A. 2017 Nov 28;114(48):12797-12802. doi: 10.1073/pnas.1705364114. Epub 2017 Nov 13. Proc Natl Acad Sci U S A. 2017. PMID: 29133396 Free PMC article.
-
Transcriptional regulation of follicular T-helper (Tfh) cells.Immunol Rev. 2013 Mar;252(1):139-45. doi: 10.1111/imr.12040. Immunol Rev. 2013. PMID: 23405901 Free PMC article. Review.
-
Differentiation of germinal center B cells and follicular helper T cells as viewed by tracking Bcl6 expression dynamics.Immunol Rev. 2012 May;247(1):120-32. doi: 10.1111/j.1600-065X.2012.01120.x. Immunol Rev. 2012. PMID: 22500836 Review.
Cited by
-
Gender-specific differences in PPARγ regulation of follicular helper T cell responses with estrogen.Sci Rep. 2016 Jun 23;6:28495. doi: 10.1038/srep28495. Sci Rep. 2016. PMID: 27335315 Free PMC article.
-
Sclerostin domain-containing protein 1 is dispensable for the differentiation of follicular helper and follicular regulatory T cells during acute viral infection.Am J Transl Res. 2019 Jun 15;11(6):3722-3736. eCollection 2019. Am J Transl Res. 2019. PMID: 31312383 Free PMC article.
-
Bcl6 controls meningeal Th17-B cell interaction in murine neuroinflammation.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2023174118. doi: 10.1073/pnas.2023174118. Proc Natl Acad Sci U S A. 2021. PMID: 34479995 Free PMC article.
-
Insights into the development and regulation of T follicular helper cells.Cytokine. 2016 Nov;87:9-19. doi: 10.1016/j.cyto.2016.06.010. Epub 2016 Jun 20. Cytokine. 2016. PMID: 27339151 Free PMC article. Review.
-
Glyphosate and AMPA Induce Alterations in Expression of Genes Involved in Chromatin Architecture in Human Peripheral Blood Mononuclear Cells (In Vitro).Int J Mol Sci. 2021 Mar 15;22(6):2966. doi: 10.3390/ijms22062966. Int J Mol Sci. 2021. PMID: 33803994 Free PMC article.
References
-
- Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, Kitagawa Y, Schneider C, Alvarez MJ, Califano A, Dalla-Favera R. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

