Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression
- PMID: 26876201
- PMCID: PMC5024153
- DOI: 10.1038/onc.2016.5
Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression
Abstract
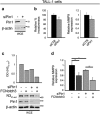
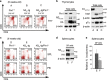
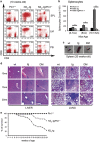
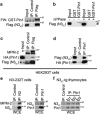
Deregulated Notch signaling is associated with T-cell Acute Lymphoblastic Leukemia (T-ALL) development and progression. Increasing evidence reveals that Notch pathway has an important role in the invasion ability of tumor cells, including leukemia, although the underlying molecular mechanisms remain mostly unclear. Here, we show that Notch3 is a novel target protein of the prolyl-isomerase Pin1, which is able to regulate Notch3 protein processing and to stabilize the cleaved product, leading to the increased expression of the intracellular domain (N3IC), finally enhancing Notch3-dependent invasiveness properties. We demonstrate that the combined inhibition of Notch3 and Pin1 in the Notch3-overexpressing human leukemic TALL-1 cells reduces their high invasive potential, by decreasing the expression of the matrix metalloprotease MMP9. Consistently, Pin1 depletion in a mouse model of Notch3-induced T-ALL, by reducing N3IC expression and signaling, impairs the expansion/invasiveness of CD4(+)CD8(+) DP cells in peripheral lymphoid and non-lymphoid organs. Notably, in in silico gene expression analysis of human T-ALL samples we observed a significant correlation between Pin1 and Notch3 expression levels, which may further suggest a key role of the newly identified Notch3-Pin1 axis in T-ALL aggressiveness and progression. Thus, combined suppression of Pin1 and Notch3 proteins may be exploited as an additional target therapy for T-ALL.
Figures


References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science 1999; 284: 770–776. - PubMed
-
- Screpanti I, Bellavia D, Campese AF, Frati L, Gulino A. Notch a unifying target in T-cell acute lymphoblastic leukemia? Trends Mol Med 2003; 9: 30–35. - PubMed
-
- Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer 2006; 6: 347–359. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

