Recruitment, Duplex Unwinding and Protein-Mediated Inhibition of the Dead-Box RNA Helicase Dbp2 at Actively Transcribed Chromatin
- PMID: 26876600
- PMCID: PMC4975007
- DOI: 10.1016/j.jmb.2016.02.005
Recruitment, Duplex Unwinding and Protein-Mediated Inhibition of the Dead-Box RNA Helicase Dbp2 at Actively Transcribed Chromatin
Abstract
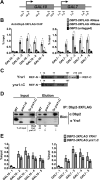
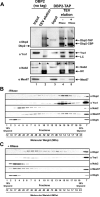
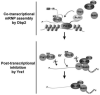
RNA helicases play fundamental roles in modulating RNA structures and facilitating RNA-protein (RNP) complex assembly in vivo. Previously, our laboratory demonstrated that the DEAD-box RNA helicase Dbp2 in Saccharomyces cerevisiae is required to promote efficient assembly of the co-transcriptionally associated mRNA-binding proteins Yra1, Nab2, and Mex67 onto poly(A)(+)RNA. We also found that Yra1 associates directly with Dbp2 and functions as an inhibitor of Dbp2-dependent duplex unwinding, suggestive of a cycle of unwinding and inhibition by Dbp2. To test this, we undertook a series of experiments to shed light on the order of events for Dbp2 in co-transcriptional mRNP assembly. We now show that Dbp2 is recruited to chromatin via RNA and forms a large, RNA-dependent complex with Yra1 and Mex67. Moreover, single-molecule fluorescence resonance energy transfer and bulk biochemical assays show that Yra1 inhibits unwinding in a concentration-dependent manner by preventing the association of Dbp2 with single-stranded RNA. This inhibition prevents over-accumulation of Dbp2 on mRNA and stabilization of a subset of RNA polymerase II transcripts. We propose a model whereby Yra1 terminates a cycle of mRNP assembly by Dbp2.
Keywords: DEAD-box; RNA; RNA–protein complex; chromatin; helicase.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
The functional complexity of the RNA-binding protein Yra1: mRNA biogenesis, genome stability and DSB repair.Curr Genet. 2020 Feb;66(1):63-71. doi: 10.1007/s00294-019-01011-8. Epub 2019 Jul 10. Curr Genet. 2020. PMID: 31292684 Review.
-
The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly.J Mol Biol. 2013 Oct 23;425(20):3824-38. doi: 10.1016/j.jmb.2013.05.016. Epub 2013 May 28. J Mol Biol. 2013. PMID: 23721653 Free PMC article.
-
Measuring helicase inhibition of the DEAD-box protein Dbp2 by Yra1.Methods Mol Biol. 2015;1259:183-97. doi: 10.1007/978-1-4939-2214-7_12. Methods Mol Biol. 2015. PMID: 25579587 Free PMC article.
-
The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription.J Biol Chem. 2012 Jul 27;287(31):26155-66. doi: 10.1074/jbc.M112.383075. Epub 2012 Jun 7. J Biol Chem. 2012. PMID: 22679025 Free PMC article.
-
The DDX5/Dbp2 subfamily of DEAD-box RNA helicases.Wiley Interdiscip Rev RNA. 2019 Mar;10(2):e1519. doi: 10.1002/wrna.1519. Epub 2018 Dec 2. Wiley Interdiscip Rev RNA. 2019. PMID: 30506978 Free PMC article. Review.
Cited by
-
DEAD-box ATPase Dbp2 is the key enzyme in an mRNP assembly checkpoint at the 3'-end of genes and involved in the recycling of cleavage factors.Nat Commun. 2024 Aug 9;15(1):6829. doi: 10.1038/s41467-024-51035-z. Nat Commun. 2024. PMID: 39122693 Free PMC article.
-
The functional complexity of the RNA-binding protein Yra1: mRNA biogenesis, genome stability and DSB repair.Curr Genet. 2020 Feb;66(1):63-71. doi: 10.1007/s00294-019-01011-8. Epub 2019 Jul 10. Curr Genet. 2020. PMID: 31292684 Review.
-
Promoter architecture determines cotranslational regulation of mRNA.Genome Res. 2018 Apr;28(4):509-518. doi: 10.1101/gr.230458.117. Epub 2018 Mar 22. Genome Res. 2018. PMID: 29567675 Free PMC article.
-
Gle1 is required for tRNA to stimulate Dbp5 ATPase activity in vitro and to promote Dbp5 mediated tRNA export in vivo.bioRxiv [Preprint]. 2023 Nov 9:2023.06.29.547072. doi: 10.1101/2023.06.29.547072. bioRxiv. 2023. Update in: Elife. 2024 Jan 08;12:RP89835. doi: 10.7554/eLife.89835. PMID: 37425677 Free PMC article. Updated. Preprint.
-
Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with S. cerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism.RNA. 2017 Jul;23(7):1125-1138. doi: 10.1261/rna.060335.116. Epub 2017 Apr 14. RNA. 2017. PMID: 28411202 Free PMC article.
References
-
- Zorio DA, Bentley DL. The link between mRNA processing and transcription: communication works both ways. Exp Cell Res. 2004;296:91–97. doi:10.1016/j.yexcr.2004.03.019S0014482704001326 [pii]. - PubMed
-
- Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, et al. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. doi:S0014-5793(01)02485-1 [pii]. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

