Single-cell Transcriptome Study as Big Data
- PMID: 26876720
- PMCID: PMC4792842
- DOI: 10.1016/j.gpb.2016.01.005
Single-cell Transcriptome Study as Big Data
Abstract
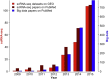
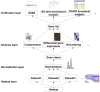
The rapid growth of single-cell RNA-seq studies (scRNA-seq) demands efficient data storage, processing, and analysis. Big-data technology provides a framework that facilitates the comprehensive discovery of biological signals from inter-institutional scRNA-seq datasets. The strategies to solve the stochastic and heterogeneous single-cell transcriptome signal are discussed in this article. After extensively reviewing the available big-data applications of next-generation sequencing (NGS)-based studies, we propose a workflow that accounts for the unique characteristics of scRNA-seq data and primary objectives of single-cell studies.
Keywords: Big data; RNA-seq; Signal normalization; Single cell; Transcriptional heterogeneity.
Copyright © 2016 The Authors. Production and hosting by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
Data Analysis in Single-Cell Transcriptome Sequencing.Methods Mol Biol. 2018;1754:311-326. doi: 10.1007/978-1-4939-7717-8_18. Methods Mol Biol. 2018. PMID: 29536451
-
High Throughput Single Cell RNA Sequencing, Bioinformatics Analysis and Applications.Adv Exp Med Biol. 2018;1068:33-43. doi: 10.1007/978-981-13-0502-3_4. Adv Exp Med Biol. 2018. PMID: 29943294 Review.
-
Single-Cell Capture, RNA-seq, and Transcriptome Analysis from the Neural Retina.Methods Mol Biol. 2020;2092:159-186. doi: 10.1007/978-1-0716-0175-4_12. Methods Mol Biol. 2020. PMID: 31786788
-
JingleBells: A Repository of Immune-Related Single-Cell RNA-Sequencing Datasets.J Immunol. 2017 May 1;198(9):3375-3379. doi: 10.4049/jimmunol.1700272. J Immunol. 2017. PMID: 28416714
-
Design and computational analysis of single-cell RNA-sequencing experiments.Genome Biol. 2016 Apr 7;17:63. doi: 10.1186/s13059-016-0927-y. Genome Biol. 2016. PMID: 27052890 Free PMC article. Review.
Cited by
-
Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment.Front Immunol. 2020 Aug 4;11:1731. doi: 10.3389/fimmu.2020.01731. eCollection 2020. Front Immunol. 2020. PMID: 32849616 Free PMC article. Review.
-
scRNA-seq for Microcephaly Research [III]: Computational Analysis of scRNA-seq Data.Methods Mol Biol. 2023;2583:105-121. doi: 10.1007/978-1-0716-2752-5_10. Methods Mol Biol. 2023. PMID: 36418729
-
The basic and translational science year in review: Confucius in the era of Big Data.Semin Arthritis Rheum. 2020 Jun;50(3):373-379. doi: 10.1016/j.semarthrit.2020.02.010. Epub 2020 Mar 5. Semin Arthritis Rheum. 2020. PMID: 32238274 Free PMC article. Review.
-
Single-cell transcriptome provides novel insights into antler stem cells, a cell type capable of mammalian organ regeneration.Funct Integr Genomics. 2019 Jul;19(4):555-564. doi: 10.1007/s10142-019-00659-2. Epub 2019 Jan 23. Funct Integr Genomics. 2019. PMID: 30673893
-
Single-cell genome-wide studies give new insight into nongenetic cell-to-cell variability in animals.Histochem Cell Biol. 2016 Sep;146(3):239-54. doi: 10.1007/s00418-016-1466-z. Epub 2016 Jul 13. Histochem Cell Biol. 2016. PMID: 27412014 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

