Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair
- PMID: 26876963
- PMCID: PMC4753642
- DOI: 10.1186/s13073-016-0273-4
Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair
Abstract
Background: Nuclease-based technologies have been developed that enable targeting of specific DNA sequences directly in the zygote. These approaches provide an opportunity to modify the genomes of inbred mice, and allow the removal of strain-specific mutations that confound phenotypic assessment. One such mutation is the Cdh23 (ahl) allele, present in several commonly used inbred mouse strains, which predisposes to age-related progressive hearing loss.
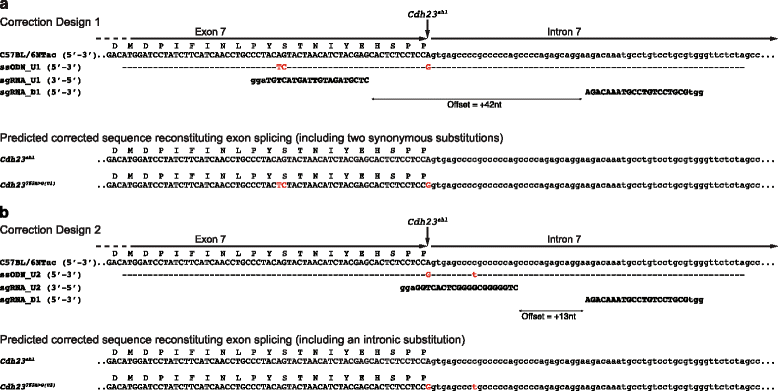
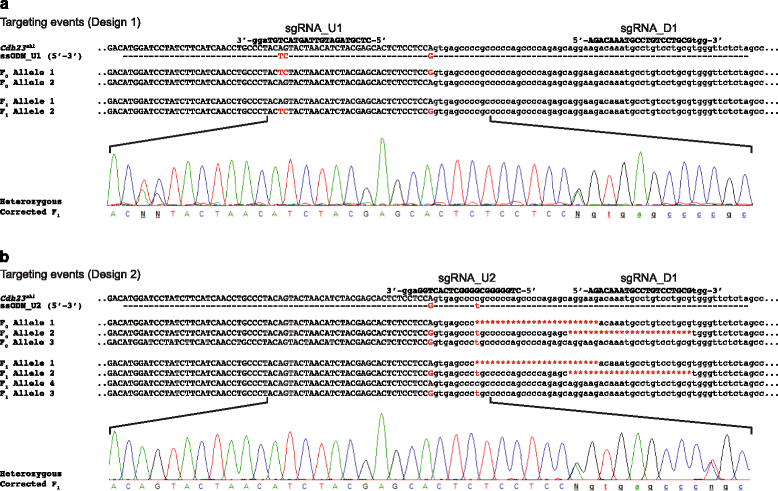
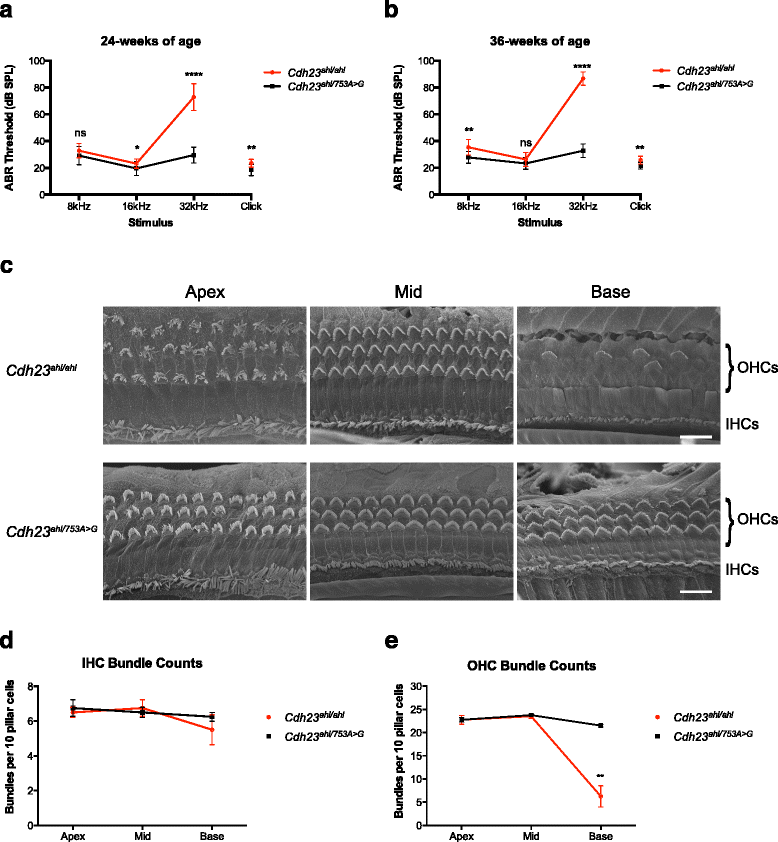
Results: We have used targeted CRISPR/Cas9-mediated homology directed repair (HDR) to correct the Cdh23 (ahl) allele directly in C57BL/6NTac zygotes. Employing offset-nicking Cas9 (D10A) nickase with paired RNA guides and a single-stranded oligonucleotide donor template we show that allele repair was successfully achieved. To investigate potential Cas9-mediated 'off-target' mutations in our corrected mouse, we undertook whole-genome sequencing and assessed the 'off-target' sites predicted for the guide RNAs (≤4 nucleotide mis-matches). No induced sequence changes were identified at any of these sites. Correction of the progressive hearing loss phenotype was demonstrated using auditory-evoked brainstem response testing of mice at 24 and 36 weeks of age, and rescue of the progressive loss of sensory hair cell stereocilia bundles was confirmed using scanning electron microscopy of dissected cochleae from 36-week-old mice.
Conclusions: CRISPR/Cas9-mediated HDR has been successfully utilised to efficiently correct the Cdh23 (ahl) allele in C57BL/6NTac mice, and rescue the associated auditory phenotype. The corrected mice described in this report will allow age-related auditory phenotyping studies to be undertaken using C57BL/6NTac-derived models, such as those generated by the International Mouse Phenotyping Consortium (IMPC) programme.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

