The presence of extra chromosomes leads to genomic instability
- PMID: 26876972
- PMCID: PMC4756715
- DOI: 10.1038/ncomms10754
The presence of extra chromosomes leads to genomic instability
Abstract
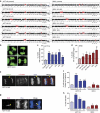
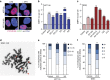
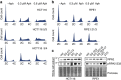
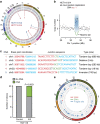
Aneuploidy is a hallmark of cancer and underlies genetic disorders characterized by severe developmental defects, yet the molecular mechanisms explaining its effects on cellular physiology remain elusive. Here we show, using a series of human cells with defined aneuploid karyotypes, that gain of a single chromosome increases genomic instability. Next-generation sequencing and SNP-array analysis reveal accumulation of chromosomal rearrangements in aneuploids, with break point junction patterns suggestive of replication defects. Trisomic and tetrasomic cells also show increased DNA damage and sensitivity to replication stress. Strikingly, we find that aneuploidy-induced genomic instability can be explained by the reduced expression of the replicative helicase MCM2-7. Accordingly, restoring near-wild-type levels of chromatin-bound MCM helicase partly rescues the genomic instability phenotypes. Thus, gain of chromosomes triggers replication stress, thereby promoting genomic instability and possibly contributing to tumorigenesis.
Figures

References
-
- Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (2016). (eds Mitelman, F., Johansson, B., Mertens, F.), http://cgap.nci.nih.gov/Chromosomes/Mitelman .
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

