Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors
- PMID: 26877223
- PMCID: PMC4779406
- DOI: 10.1016/j.stem.2015.12.001
Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors
Abstract
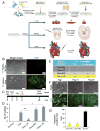
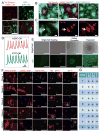
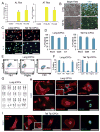
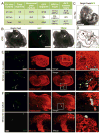
Several studies have reported reprogramming of fibroblasts into induced cardiomyocytes; however, reprogramming into proliferative induced cardiac progenitor cells (iCPCs) remains to be accomplished. Here we report that a combination of 11 or 5 cardiac factors along with canonical Wnt and JAK/STAT signaling reprogrammed adult mouse cardiac, lung, and tail tip fibroblasts into iCPCs. The iCPCs were cardiac mesoderm-restricted progenitors that could be expanded extensively while maintaining multipotency to differentiate into cardiomyocytes, smooth muscle cells, and endothelial cells in vitro. Moreover, iCPCs injected into the cardiac crescent of mouse embryos differentiated into cardiomyocytes. iCPCs transplanted into the post-myocardial infarction mouse heart improved survival and differentiated into cardiomyocytes, smooth muscle cells, and endothelial cells. Lineage reprogramming of adult somatic cells into iCPCs provides a scalable cell source for drug discovery, disease modeling, and cardiac regenerative therapy.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Are we closer to cardiac regeneration?Stem Cell Investig. 2016 Oct 20;3:59. doi: 10.21037/sci.2016.09.16. eCollection 2016. Stem Cell Investig. 2016. PMID: 27868041 Free PMC article. No abstract available.
-
Novel direct reprogramming technique for the generation of culture-expandable cardiac progenitor cells from fibroblasts.Stem Cell Investig. 2017 Feb 16;4:15. doi: 10.21037/sci.2017.02.04. eCollection 2017. Stem Cell Investig. 2017. PMID: 28275645 Free PMC article. No abstract available.
Similar articles
-
The Future of Direct Cardiac Reprogramming: Any GMT Cocktail Variety?Int J Mol Sci. 2020 Oct 26;21(21):7950. doi: 10.3390/ijms21217950. Int J Mol Sci. 2020. PMID: 33114756 Free PMC article. Review.
-
Induced cardiac progenitor cells repopulate decellularized mouse heart scaffolds and differentiate to generate cardiac tissue.Biochim Biophys Acta Mol Cell Res. 2020 Mar;1867(3):118559. doi: 10.1016/j.bbamcr.2019.118559. Epub 2019 Oct 18. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 31634503 Free PMC article.
-
Generation of multipotent induced cardiac progenitor cells from mouse fibroblasts and potency testing in ex vivo mouse embryos.Nat Protoc. 2017 May;12(5):1029-1054. doi: 10.1038/nprot.2017.021. Epub 2017 Apr 20. Nat Protoc. 2017. PMID: 28426026 Free PMC article.
-
Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts.Cell Stem Cell. 2016 Mar 3;18(3):368-81. doi: 10.1016/j.stem.2016.02.001. Cell Stem Cell. 2016. PMID: 26942852 Free PMC article.
-
Generation of induced cardiac progenitor cells via somatic reprogramming.Oncotarget. 2017 Apr 25;8(17):29442-29457. doi: 10.18632/oncotarget.15272. Oncotarget. 2017. PMID: 28199972 Free PMC article. Review.
Cited by
-
Improving cardiac reprogramming for heart regeneration.Curr Opin Organ Transplant. 2016 Dec;21(6):588-594. doi: 10.1097/MOT.0000000000000363. Curr Opin Organ Transplant. 2016. PMID: 27755167 Free PMC article. Review.
-
Cardiac Progenitor Cells from Stem Cells: Learning from Genetics and Biomaterials.Cells. 2019 Nov 28;8(12):1536. doi: 10.3390/cells8121536. Cells. 2019. PMID: 31795206 Free PMC article. Review.
-
Generation of NKX2.5GFP Reporter Human iPSCs and Differentiation Into Functional Cardiac Fibroblasts.Front Cell Dev Biol. 2022 Jan 21;9:797927. doi: 10.3389/fcell.2021.797927. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35127713 Free PMC article.
-
Cardiac progenitor reprogramming for heart regeneration.Cell Regen. 2018 Feb 12;7(1):1-6. doi: 10.1016/j.cr.2018.01.001. eCollection 2018 Sep. Cell Regen. 2018. PMID: 30671223 Free PMC article. Review.
-
The Art of Reprogramming for Regenerative Medicine.Front Cell Dev Biol. 2022 Jun 30;10:927555. doi: 10.3389/fcell.2022.927555. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35846373 Free PMC article. Review.
References
-
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annual review of cell and developmental biology. 2007;23:45–68. - PubMed
-
- Birket MJ, Ribeiro MC, Verkerk AO, Ward D, Leitoguinho AR, den Hartogh SC, Orlova VV, Devalla HD, Schwach V, Bellin M, et al. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nature biotechnology. 2015;33:970–979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

