Selective and Potent Proteomimetic Inhibitors of Intracellular Protein-Protein Interactions
- PMID: 26877561
- PMCID: PMC4737265
- DOI: 10.1002/ange.201410810
Selective and Potent Proteomimetic Inhibitors of Intracellular Protein-Protein Interactions
Abstract
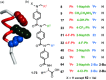
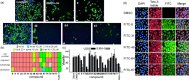
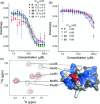
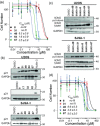
Inhibition of protein-protein interactions (PPIs) represents a major challenge in chemical biology and drug discovery. α-Helix mediated PPIs may be amenable to modulation using generic chemotypes, termed "proteomimetics", which can be assembled in a modular manner to reproduce the vectoral presentation of key side chains found on a helical motif from one partner within the PPI. In this work, it is demonstrated that by using a library of N-alkylated aromatic oligoamide helix mimetics, potent helix mimetics which reproduce their biophysical binding selectivity in a cellular context can be identified.
Wählerische Mimetika: Die Inhibierung von Protein‐Protein‐Wechselwirkungen ist eine zentrale Aufgabe in der chemischen Biologie sowie in der Entdeckung und Entwicklung neuer Wirkstoffe. Anhand einer Bibliothek von N‐alkylierten aromatischen Oligoamiden wird gezeigt, dass Helixmimetika identifiziert werden können, die ihre biophysikalische Bindungsselektivität in einem zellulären Umfeld reproduzieren.WILEY-VCH.
Keywords: Apoptose; Foldamere; Helikale Strukturen; Peptidomimetika; Protein‐Protein‐Wechselwirkungen.
Figures

References
-
- Milroy L.‐G., Grossmann T. N., Hennig S., Brunsveld L., Ottmann C., Chem. Rev. 2014, 114, 4695. - PubMed
-
- Berg T., Angew. Chem. Int. Ed. 2003, 42, 2462; - PubMed
- Angew. Chem. 2003, 115, 2566.
-
- Yin H., Hamilton A. D., Angew. Chem. Int. Ed. 2005, 44, 4130; - PubMed
- Angew. Chem. 2005, 117, 4200.
-
- Wells J. A., McLendon C. L., Nature 2007, 450, 1001. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
