Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers
- PMID: 26878173
- PMCID: PMC4767360
- DOI: 10.1172/JCI85271
Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers
Abstract
Background: Poorly differentiated thyroid cancer (PDTC) and anaplastic thyroid cancer (ATC) are rare and frequently lethal tumors that so far have not been subjected to comprehensive genetic characterization.
Methods: We performed next-generation sequencing of 341 cancer genes from 117 patient-derived PDTCs and ATCs and analyzed the transcriptome of a representative subset of 37 tumors. Results were analyzed in the context of The Cancer Genome Atlas study (TCGA study) of papillary thyroid cancers (PTC).
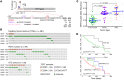
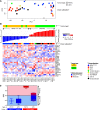
Results: Compared to PDTCs, ATCs had a greater mutation burden, including a higher frequency of mutations in TP53, TERT promoter, PI3K/AKT/mTOR pathway effectors, SWI/SNF subunits, and histone methyltransferases. BRAF and RAS were the predominant drivers and dictated distinct tropism for nodal versus distant metastases in PDTC. RAS and BRAF sharply distinguished between PDTCs defined by the Turin (PDTC-Turin) versus MSKCC (PDTC-MSK) criteria, respectively. Mutations of EIF1AX, a component of the translational preinitiation complex, were markedly enriched in PDTCs and ATCs and had a striking pattern of co-occurrence with RAS mutations. While TERT promoter mutations were rare and subclonal in PTCs, they were clonal and highly prevalent in advanced cancers. Application of the TCGA-derived BRAF-RAS score (a measure of MAPK transcriptional output) revealed a preserved relationship with BRAF/RAS mutation in PDTCs, whereas ATCs were BRAF-like irrespective of driver mutation.
Conclusions: These data support a model of tumorigenesis whereby PDTCs and ATCs arise from well-differentiated tumors through the accumulation of key additional genetic abnormalities, many of which have prognostic and possible therapeutic relevance. The widespread genomic disruptions in ATC compared with PDTC underscore their greater virulence and higher mortality.
Funding: This work was supported in part by NIH grants CA50706, CA72597, P50-CA72012, P30-CA008748, and 5T32-CA160001; the Lefkovsky Family Foundation; the Society of Memorial Sloan Kettering; the Byrne fund; and Cycle for Survival.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

