Statistical correlation of nonconservative substitutions of HIV gp41 variable amino acid residues with the R5X4 HIV-1 phenotype
- PMID: 26879054
- PMCID: PMC4754869
- DOI: 10.1186/s12985-016-0486-6
Statistical correlation of nonconservative substitutions of HIV gp41 variable amino acid residues with the R5X4 HIV-1 phenotype
Abstract
Background: The interaction of the envelope glycoprotein of HIV-1 (gp120/gp41) with coreceptor molecules has important implications for specific cellular targeting and pathogenesis. Experimental and theoretical evidences have shown a role for gp41 in coreceptor tropism, although there is no consensus about the positions involved. Here we analyze the association of physicochemical properties of gp41 amino acid residues with viral tropism (X4, R5, and R5X4) using a large set of HIV-1 sequences. Under the assumption that conserved regions define the complex structural features essential for protein function, we focused our search only on amino acids in the gp41 variable regions.
Methods: Gp41 amino acid sequences of 2823 HIV-1 strains from all clades with known coreceptor tropism were retrieved from Los Alamos HIV Database. Consensus sequences were constructed for homologous sequences (those obtained from the same patient and having the same tropism) in order to avoid bias due to sequence overrepresentation, and the variability (entropy) per site was determined. Comparisons of hydropathy index (HI) and charge (Q) of amino acid residues at highly variable positions between coreceptor groups were performed using two non-parametrical tests and Benjamini-Hochberg correction. Pearson's correlation analysis was performed to determine covariance of HI and Q values.
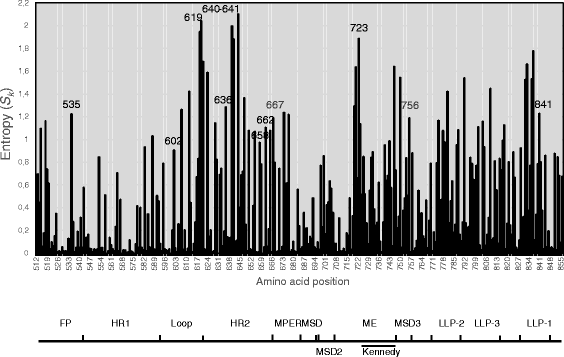
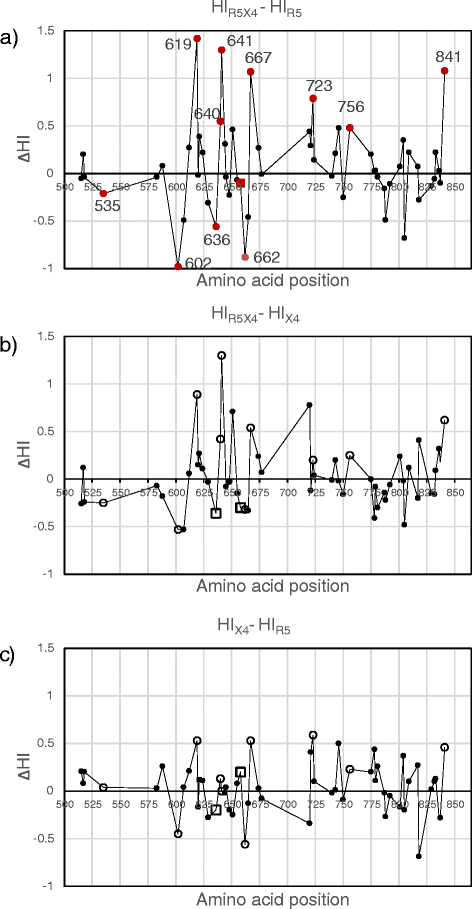
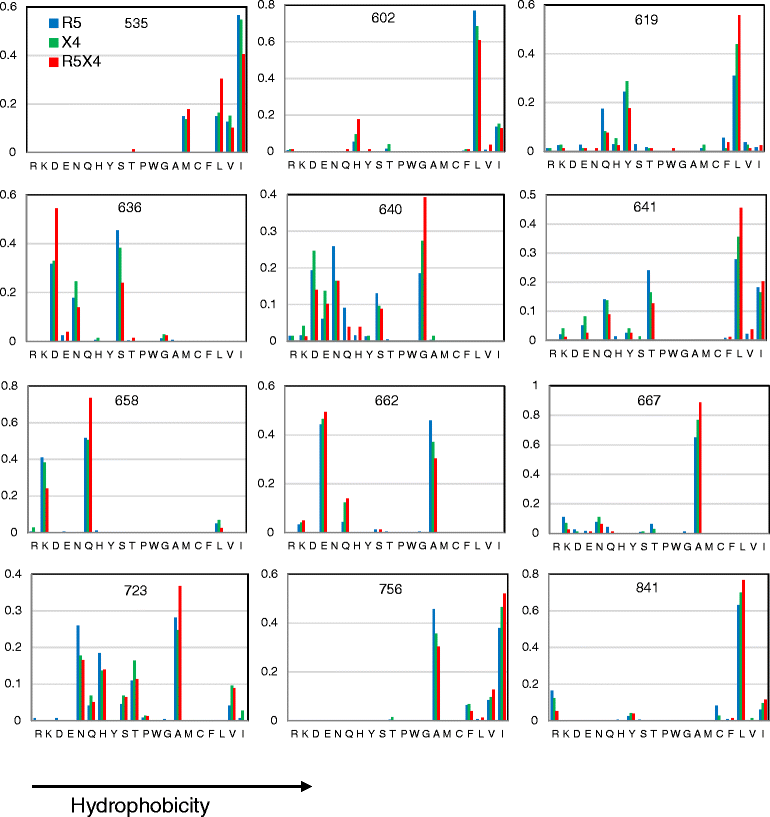
Results: Calculation of variability per site rendered 58 highly variable amino acid positions. Of these, statistical analysis rendered significantly different HI or Q only for the R5 vs. R5X4 comparison at twelve positions: 535, 602, 619, 636, 640, 641, 658, 662, 667, 723, 756 and 841. The largest differences in particular amino acid frequencies between coreceptor groups were found at 619, 636, 640, 641, 662, 723 and 756. A hydrophobic tendency of residues 619, 640, 641, 723 and 756, along with a hydrophilic/charged tendency at residues 636 and 662 was observed in R5X4 with respect to R5 sequences. HI of position 640 covariated with that of 602, 619, 636, 662, and 756.
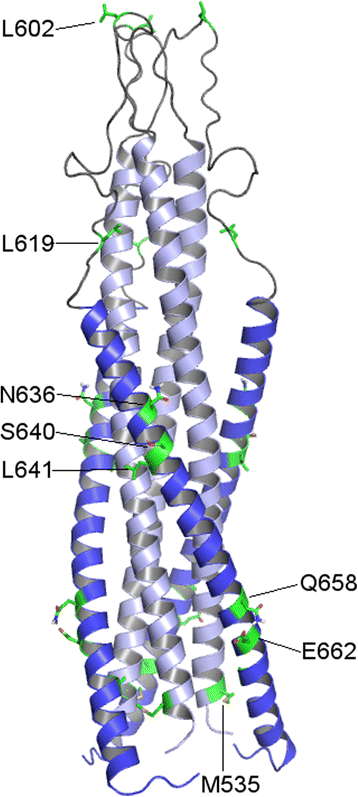
Conclusions: Variability and significant correlations of physicochemical properties with viral phenotype suggest that substitutions at residues in the loop (602 and 619), the HR2 (636, 640, 641, 662), and the C-terminal tail (723, 756) of gp41 may contribute to phenotype of R5X4 strains.
Figures
Similar articles
-
On the Physicochemical and Structural Modifications Associated with HIV-1 Subtype B Tropism Transition.AIDS Res Hum Retroviruses. 2016 Aug;32(8):829-40. doi: 10.1089/AID.2015.0373. Epub 2016 Jun 1. AIDS Res Hum Retroviruses. 2016. PMID: 27071630 Free PMC article.
-
Selected amino acid changes in HIV-1 subtype-C gp41 are associated with specific gp120(V3) signatures in the regulation of co-receptor usage.Virus Res. 2012 Sep;168(1-2):73-83. doi: 10.1016/j.virusres.2012.06.019. Epub 2012 Jun 23. Virus Res. 2012. PMID: 22732432
-
Selected amino acid mutations in HIV-1 B subtype gp41 are associated with specific gp120v₃ signatures in the regulation of co-receptor usage.Retrovirology. 2011 May 12;8:33. doi: 10.1186/1742-4690-8-33. Retrovirology. 2011. PMID: 21569409 Free PMC article.
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
-
N- and C-domains of HIV-1 gp41: mutation, structure and functions.Immunol Lett. 2001 Jan 15;75(3):215-20. doi: 10.1016/s0165-2478(00)00302-3. Immunol Lett. 2001. PMID: 11166378 Review.
Cited by
-
Mutational networks of escape from transmitted HIV-1 infection.PLoS One. 2020 Dec 7;15(12):e0243391. doi: 10.1371/journal.pone.0243391. eCollection 2020. PLoS One. 2020. PMID: 33284837 Free PMC article.
-
Accurate predictions of population-level changes in sequence and structural properties of HIV-1 Env using a volatility-controlled diffusion model.PLoS Biol. 2017 Apr 6;15(4):e2001549. doi: 10.1371/journal.pbio.2001549. eCollection 2017 Apr. PLoS Biol. 2017. PMID: 28384158 Free PMC article.
-
Analysis of sequence diversity and selection pressure in HIV-1 clade C gp41 from India.Virusdisease. 2020 Sep;31(3):277-291. doi: 10.1007/s13337-020-00595-x. Epub 2020 May 12. Virusdisease. 2020. PMID: 32904888 Free PMC article.
References
-
- Steckbeck JD, Craigo JK, Barnes CO, Montelaro RC. Highly conserved structural properties of the C-terminal tail of HIV-1 gp41 protein despite substantial sequence variation among diverse clades: implications for functions in viral replication. J Biol Chem. 2011;286(31):27156–66. doi: 10.1074/jbc.M111.258855. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

