The Genetic History of Peruvian Quechua-Lamistas and Chankas: Uniparental DNA Patterns among Autochthonous Amazonian and Andean Populations
- PMID: 26879156
- PMCID: PMC5111738
- DOI: 10.1111/ahg.12145
The Genetic History of Peruvian Quechua-Lamistas and Chankas: Uniparental DNA Patterns among Autochthonous Amazonian and Andean Populations
Abstract
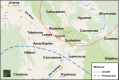
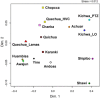
This study focuses on the genetic history of the Quechua-Lamistas, inhabitants of the Lamas Province in the San Martin Department, Peru, who speak their own distinct variety of the Quechua family of languages. It has been suggested that different pre-Columbian ethnic groups from the Peruvian Amazonia, like the Motilones or "shaven heads", assimilated the Quechua language and then formed the current native population of Lamas. However, many Quechua-Lamistas claim to be direct descendants of the Chankas, a famous pre-Columbian indigenous group that escaped from Inca rule in the Andes. To investigate the Quechua-Lamistas and Chankas' ancestries, we compared uniparental genetic profiles (17 STRs of Q-M3 Y-chromosome and mtDNA complete control region haplotypes) among autochthonous Amazonian and Andean populations from Peru, Bolivia and Ecuador. The phylogeographic and population genetic analyses indicate a fairly heterogeneous ancestry for the Quechua-Lamistas, while they are closely related to their neighbours who speak Amazonian languages, presenting no direct relationships with populations from the region where the ancient Chankas lived. On the other hand, the genetic profiles of self-identified Chanka descendants living in Andahuaylas (located in the Apurimac Department, Peru, in the Central Andes) were closely related to those living in Huancavelica and the assumed Chanka Confederation area before the Inca expansion.
Keywords: Amazonia; Andes; Chankas; Quechua-Lamistas; Y-SNPs; Y-STRs; human history; indigenous South Americans; mtDNA.
© 2016 John Wiley & Sons Ltd/University College London.
Figures



Similar articles
-
The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: the legacy of the Uros.PLoS One. 2013 Sep 11;8(9):e73006. doi: 10.1371/journal.pone.0073006. eCollection 2013. PLoS One. 2013. PMID: 24039843 Free PMC article.
-
Tracing the genetic history of the 'Cañaris' from Ecuador and Peru using uniparental DNA markers.BMC Genomics. 2020 Sep 10;21(Suppl 7):413. doi: 10.1186/s12864-020-06834-1. BMC Genomics. 2020. PMID: 32912150 Free PMC article.
-
Population genetic structure of traditional populations in the Peruvian Central Andes and implications for South American population history.Hum Biol. 2014 Summer;86(3):147-65. doi: 10.13110/humanbiology.86.3.0147. Hum Biol. 2014. PMID: 25836744
-
Genetic approaches to understanding human adaptation to altitude in the Andes.J Exp Biol. 2001 Sep;204(Pt 18):3151-60. doi: 10.1242/jeb.204.18.3151. J Exp Biol. 2001. PMID: 11581329 Review.
-
The home team advantage: reproduction in women indigenous to high altitude.J Exp Biol. 2001 Sep;204(Pt 18):3141-50. doi: 10.1242/jeb.204.18.3141. J Exp Biol. 2001. PMID: 11581328 Review.
Cited by
-
Uniparental Lineages from the Oldest Indigenous Population of Ecuador: The Tsachilas.Genes (Basel). 2021 Aug 20;12(8):1273. doi: 10.3390/genes12081273. Genes (Basel). 2021. PMID: 34440446 Free PMC article.
-
Hepatitis C virus pharmacogenomics in Latin American populations: implications in the era of direct-acting antivirals.Pharmgenomics Pers Med. 2017 Mar 28;10:79-91. doi: 10.2147/PGPM.S125452. eCollection 2017. Pharmgenomics Pers Med. 2017. PMID: 28405170 Free PMC article. Review.
-
The Current Genomic Landscape of Western South America: Andes, Amazonia, and Pacific Coast.Mol Biol Evol. 2019 Dec 1;36(12):2698-2713. doi: 10.1093/molbev/msz174. Mol Biol Evol. 2019. PMID: 31350885 Free PMC article.
-
Genetic ancestry of families of putative Inka descent.Mol Genet Genomics. 2018 Aug;293(4):873-881. doi: 10.1007/s00438-018-1427-4. Epub 2018 Mar 3. Mol Genet Genomics. 2018. PMID: 29502256 Free PMC article. Clinical Trial.
-
Maternal ancestry and lineages diversity of the Santander population from Colombia.Forensic Sci Res. 2023 Sep 21;8(3):241-248. doi: 10.1093/fsr/owad032. eCollection 2023 Sep. Forensic Sci Res. 2023. PMID: 38221971 Free PMC article.
References
-
- Adelaar, W. F. H. (2013) Quechua I y Quechua II: En defensa de una distinción establecida. Rev Bras Linguist Antropol 5(1), 45–65.
-
- Andrews, R. M. , Kubacka. I. , Chinnery, P. F. , Lightowlers, R. N. , Turnbull, D. M. & Howell, N. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23, 147. - PubMed
-
- Arnaiz‐Villena, A. , Moscoso, J. , Serrano‐Vela, J. I. & Martinez‐Laso, J. (2006) The uniqueness of Amerindians according to HLA genes and the peopling of the Americas. Immunología 25, 13–24.
-
- Baeta, M. , Núñez, C. , Sosa, C. , Bolea, M. , Casalod, Y. , Gonzáles‐Andrade, F. , Roewer, L. & Martínez‐Jarreta, B. (2012) Mitochondrial diversity in Amerindian Kichwa and Mestizo populations from Ecuador. Int J Legal Med 126, 299–302. - PubMed
-
- Bandelt, H. ‐J. , Forster, P. & Röhl, A. (1999) Median‐Joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16, 37–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

