Whole genome resequencing of the human parasite Schistosoma mansoni reveals population history and effects of selection
- PMID: 26879532
- PMCID: PMC4754680
- DOI: 10.1038/srep20954
Whole genome resequencing of the human parasite Schistosoma mansoni reveals population history and effects of selection
Abstract
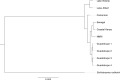
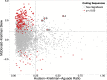
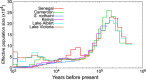
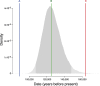
Schistosoma mansoni is a parasitic fluke that infects millions of people in the developing world. This study presents the first application of population genomics to S. mansoni based on high-coverage resequencing data from 10 global isolates and an isolate of the closely-related Schistosoma rodhaini, which infects rodents. Using population genetic tests, we document genes under directional and balancing selection in S. mansoni that may facilitate adaptation to the human host. Coalescence modeling reveals the speciation of S. mansoni and S. rodhaini as 107.5-147.6KYA, a period which overlaps with the earliest archaeological evidence for fishing in Africa. Our results indicate that S. mansoni originated in East Africa and experienced a decline in effective population size 20-90KYA, before dispersing across the continent during the Holocene. In addition, we find strong evidence that S. mansoni migrated to the New World with the 16-19th Century Atlantic Slave Trade.
Figures


References
-
- Brunet L. R., Dunne D. & Pearce E. Cytokine interaction and immune responses during Schistosoma mansoni infection. Parasitology today 14, 422–427 (1998). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

