A novel role for Crp in controlling magnetosome biosynthesis in Magnetospirillum gryphiswaldense MSR-1
- PMID: 26879571
- PMCID: PMC4754748
- DOI: 10.1038/srep21156
A novel role for Crp in controlling magnetosome biosynthesis in Magnetospirillum gryphiswaldense MSR-1
Abstract
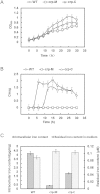
Magnetotactic bacteria (MTB) are specialized microorganisms that synthesize intracellular magnetite particles called magnetosomes. Although many studies have focused on the mechanism of magnetosome synthesis, it remains unclear how these structures are formed. Recent reports have suggested that magnetosome formation is energy dependent. To investigate the relationship between magnetosome formation and energy metabolism, a global regulator, named Crp, which mainly controls energy and carbon metabolism in most microorganisms, was genetically disrupted in Magnetospirillum gryphiswaldense MSR-1. Compared with the wild-type or complemented strains, the growth, ferromagnetism and intracellular iron content of crp-deficient mutant cells were dramatically decreased. Transmission electron microscopy (TEM) showed that magnetosome synthesis was strongly impaired by the disruption of crp. Further gene expression profile analysis showed that the disruption of crp not only influenced genes related to energy and carbon metabolism, but a series of crucial magnetosome island (MAI) genes were also down regulated. These results indicate that Crp is essential for magnetosome formation in MSR-1. This is the first time to demonstrate that Crp plays an important role in controlling magnetosome biomineralization and provides reliable expression profile data that elucidate the mechanism of Crp regulation of magnetosome formation in MSR-1.
Figures




Similar articles
-
Expression patterns of key iron and oxygen metabolism genes during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1.FEMS Microbiol Lett. 2013 Oct;347(2):163-72. doi: 10.1111/1574-6968.12234. Epub 2013 Sep 6. FEMS Microbiol Lett. 2013. PMID: 23937222
-
FeoB2 Functions in magnetosome formation and oxidative stress protection in Magnetospirillum gryphiswaldense strain MSR-1.J Bacteriol. 2012 Aug;194(15):3972-6. doi: 10.1128/JB.00382-12. Epub 2012 May 25. J Bacteriol. 2012. PMID: 22636767 Free PMC article.
-
MamX encoded by the mamXY operon is involved in control of magnetosome maturation in Magnetospirillum gryphiswaldense MSR-1.BMC Microbiol. 2013 Sep 11;13:203. doi: 10.1186/1471-2180-13-203. BMC Microbiol. 2013. PMID: 24020498 Free PMC article.
-
Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense.Arch Microbiol. 2004 Jan;181(1):1-7. doi: 10.1007/s00203-003-0631-7. Epub 2003 Dec 11. Arch Microbiol. 2004. PMID: 14668979 Review.
-
Genetics and cell biology of magnetosome formation in magnetotactic bacteria.FEMS Microbiol Rev. 2008 Jul;32(4):654-72. doi: 10.1111/j.1574-6976.2008.00116.x. Epub 2008 Jun 2. FEMS Microbiol Rev. 2008. PMID: 18537832 Review.
Cited by
-
Magnetic genes: Studying the genetics of biomineralization in magnetotactic bacteria.PLoS Genet. 2020 Feb 13;16(2):e1008499. doi: 10.1371/journal.pgen.1008499. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32053597 Free PMC article. Review.
-
The Disruption of an OxyR-Like Protein Impairs Intracellular Magnetite Biomineralization in Magnetospirillum gryphiswaldense MSR-1.Front Microbiol. 2017 Feb 14;8:208. doi: 10.3389/fmicb.2017.00208. eCollection 2017. Front Microbiol. 2017. PMID: 28261169 Free PMC article.
-
Metabolic characterisation of Magnetospirillum gryphiswaldense MSR-1 using LC-MS-based metabolite profiling.RSC Adv. 2020 Sep 2;10(54):32548-32560. doi: 10.1039/d0ra05326k. eCollection 2020 Sep 1. RSC Adv. 2020. PMID: 35516490 Free PMC article.
-
Genome-Wide Identification of Essential and Auxiliary Gene Sets for Magnetosome Biosynthesis in Magnetospirillum gryphiswaldense.mSystems. 2020 Nov 17;5(6):e00565-20. doi: 10.1128/mSystems.00565-20. mSystems. 2020. PMID: 33203687 Free PMC article.
-
Continuous Production of Biogenic Magnetite Nanoparticles by the Marine Bacterium Magnetovibrio blakemorei Strain MV-1T with a Nitrous Oxide Injection Strategy.Mar Drugs. 2022 Nov 18;20(11):724. doi: 10.3390/md20110724. Mar Drugs. 2022. PMID: 36422002 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

