Inferences from tip-calibrated phylogenies: a review and a practical guide
- PMID: 26880113
- PMCID: PMC4949988
- DOI: 10.1111/mec.13586
Inferences from tip-calibrated phylogenies: a review and a practical guide
Abstract
Molecular dating of phylogenetic trees is a growing discipline using sequence data to co-estimate the timing of evolutionary events and rates of molecular evolution. All molecular-dating methods require converting genetic divergence between sequences into absolute time. Historically, this could only be achieved by associating externally derived dates obtained from fossil or biogeographical evidence to internal nodes of the tree. In some cases, notably for fast-evolving genomes such as viruses and some bacteria, the time span over which samples were collected may cover a significant proportion of the time since they last shared a common ancestor. This situation allows phylogenetic trees to be calibrated by associating sampling dates directly to the sequences representing the tips (terminal nodes) of the tree. The increasing availability of genomic data from ancient DNA extends the applicability of such tip-based calibration to a variety of taxa including humans, extinct megafauna and various microorganisms which typically have a scarce fossil record. The development of statistical models accounting for heterogeneity in different aspects of the evolutionary process while accommodating very large data sets (e.g. whole genomes) has allowed using tip-dating methods to reach inferences on divergence times, substitution rates, past demography or the age of specific mutations on a variety of spatiotemporal scales. In this review, we summarize the current state of the art of tip dating, discuss some recent applications, highlight common pitfalls and provide a 'how to' guide to thoroughly perform such analyses.
Keywords: Bayesian phylogenetics; calibration, divergence time and substitution rate inferences; measurably evolving populations; population dynamics; tip-dating.
© 2016 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures
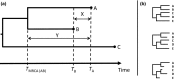

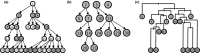
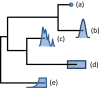

Comment in
-
Comment on Rieux and Balloux: calibration from tip-dating can compromise topological accuracy and evolutionary inference.Mol Ecol. 2017 May;26(10):2623-2624. doi: 10.1111/mec.13951. Epub 2016 Dec 31. Mol Ecol. 2017. PMID: 28039963
References
-
- Alter SE, Meyer M, Post K et al (2015) Climate impacts on transocean dispersal and habitat in gray whales from the Pleistocene to 2100. Molecular Ecology, 24, 1510–1522. - PubMed
-
- Arcila D, Pyron RA, Tyler JC, Orti G, Betancur‐R R (2015) An evaluation of fossil tip‐dating versus node‐age calibrations in tetraodontiform fishes (Teleostei: Percomorphaceae). Molecular Phylogenetics and Evolution, 82, 131–145. - PubMed
-
- Axelsson E, Willerslev E, Gilbert MTP, Nielsen R (2008) The effect of ancient DNA damage on inferences of demographic histories. Molecular Biology and Evolution, 25, 2181–2187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

