IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity
- PMID: 26880249
- PMCID: PMC4754645
- DOI: 10.1038/srep20842
IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity
Erratum in
-
Erratum: IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity.Sci Rep. 2016 Apr 29;6:23876. doi: 10.1038/srep23876. Sci Rep. 2016. PMID: 27125899 Free PMC article. No abstract available.
Abstract
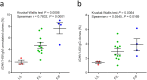
IGHV polymorphism provides a rich source of humoral immune system diversity. One important example is the IGHV1-69 germline gene where the biased use of alleles that encode the critical CDR-H2 Phe54 (F-alleles) to make broadly neutralizing antibodies (HV1-69-sBnAb) to the influenza A hemagglutinin stem domain has been clearly established. However, whether IGHV1-69 polymorphism can also modulate B cell function and Ab repertoire expression through promoter and copy number (CN) variations has not been reported, nor has whether IGHV1-69 allelic distribution is impacted by ethnicity. Here we studied a cohort of NIH H5N1 vaccinees and demonstrate for the first time the influence of IGHV1-69 polymorphism on V-segment usage, somatic hypermutation and B cell expansion that elucidates the dominance of F-alleles in HV1-69-sBnAbs. We provide evidence that Phe54/Leu54 (F/L) polymorphism correlates with shifted repertoire usage of other IGHV germline genes. In addition, we analyzed ethnically diverse individuals within the 1000 genomes project and discovered marked variations in F- and L- genotypes and CN among the various ethnic groups that may impact HV1-69-sBnAb responses. These results have immediate implications for understanding HV1-69-sBnAb responses at the individual and population level and for the design and implementation of "universal" influenza vaccine.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

